6PKX
 
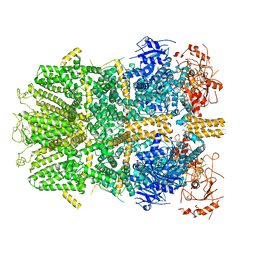 | | Cryo-EM structure of the zebrafish TRPM2 channel in the presence of ADPR and Ca2+ | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily M member 2, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Yin, Y, Wu, M, Hsu, A.L, Borschel, W.F, Borgnia, M.J, Lander, G.C, Lee, S.-Y. | | Deposit date: | 2019-06-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Visualizing structural transitions of ligand-dependent gating of the TRPM2 channel.
Nat Commun, 10, 2019
|
|
6OT5
 
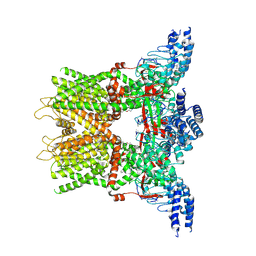 | | Structure of the TRPV3 K169A sensitized mutant in the presence of 2-APB at 3.6 A resolution | | Descriptor: | 2-aminoethyl diphenylborinate, Transient receptor potential cation channel subfamily V member 3,Transient receptor potential cation channel subfamily V member 3 | | Authors: | Zubcevic, L, Borschel, W.F, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-05-02 | | Release date: | 2019-05-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Regulatory switch at the cytoplasmic interface controls TRPV channel gating.
Elife, 8, 2019
|
|
6OUG
 
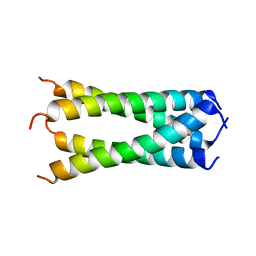 | | Structure of drug-resistant V27A mutant of the influenza M2 proton channel bound to spiroadamantyl amine inhibitor, TM + cytosolic helix construct | | Descriptor: | (1r,1'S,3'S,5'S,7'S)-spiro[cyclohexane-1,2'-tricyclo[3.3.1.1~3,7~]decan]-4-amine, Matrix protein 2 | | Authors: | Thomaston, J.L, Liu, L, DeGrado, W.F. | | Deposit date: | 2019-05-04 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | X-ray Crystal Structures of the Influenza M2 Proton Channel Drug-Resistant V27A Mutant Bound to a Spiro-Adamantyl Amine Inhibitor Reveal the Mechanism of Adamantane Resistance.
Biochemistry, 59, 2020
|
|
6OT2
 
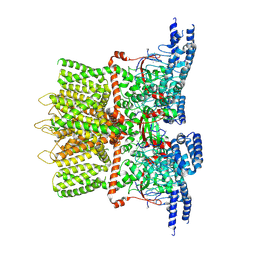 | | Structure of the TRPV3 K169A sensitized mutant in apo form at 4.1 A resolution | | Descriptor: | Transient receptor potential cation channel subfamily V member 3 | | Authors: | Zubcevic, L, Borschel, W.F, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-05-02 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Regulatory switch at the cytoplasmic interface controls TRPV channel gating.
Elife, 8, 2019
|
|
6US9
 
 | |
6US8
 
 | |
6S3N
 
 | | Crystal structure of helicase Pif1 from Thermus oshimai in complex with ssDNA (dT)18 and ADP-VO4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Dai, Y.X, Chen, W.F, Teng, F.Y, Liu, N.N, Hou, X.M, Dou, S.X, Rety, S, Xi, X.G. | | Deposit date: | 2019-06-25 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.533 Å) | | Cite: | Structural and functional studies of SF1B Pif1 from Thermus oshimai reveal dimerization-induced helicase inhibition.
Nucleic Acids Res., 49, 2021
|
|
8BFS
 
 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) in complex with FZ326 | | Descriptor: | 1,2-ETHANEDIOL, 3~{H}-pyrrolo[2,3-c]isoquinolin-5-amine, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Zhu, W.F, Hernandez-Olmos, V, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-26 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) in complex with FZ326
To Be Published
|
|
6S3I
 
 | | Crystal structure of helicase Pif1 from Thermus oshimai in complex with ssDNA (dT)18 and ADP-MgF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Dai, Y.X, Chen, W.F, Teng, F.Y, Liu, N.N, Hou, X.M, Dou, S.X, Rety, S, Xi, X.G. | | Deposit date: | 2019-06-25 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | Structural and functional studies of SF1B Pif1 from Thermus oshimai reveal dimerization-induced helicase inhibition.
Nucleic Acids Res., 49, 2021
|
|
6S3P
 
 | | Crystal structure of helicase Pif1 from Thermus oshimai in complex with (dT)18 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*T)-3'), PIF1 helicase | | Authors: | Dai, Y.X, Chen, W.F, Teng, F.Y, Liu, N.N, Hou, X.M, Dou, S.X, Rety, S, Xi, X.G. | | Deposit date: | 2019-06-25 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.926 Å) | | Cite: | Structural and functional studies of SF1B Pif1 from Thermus oshimai reveal dimerization-induced helicase inhibition.
Nucleic Acids Res., 49, 2021
|
|
8BH8
 
 | |
8BH9
 
 | |
1U7M
 
 | | Solution structure of a diiron protein model: Due Ferri(II) turn mutant | | Descriptor: | Four-helix bundle model, ZINC ION | | Authors: | Maglio, O, Nastri, F, Calhoun, J.R, Lahr, S, Pavone, V, DeGrado, W.F, Lombardi, A. | | Deposit date: | 2004-08-04 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Analysis and Design of Turns in alpha-Helical Hairpins
J.Mol.Biol., 346, 2005
|
|
6S3H
 
 | | Crystal structure of helicase Pif1 from Thermus oshimai in complex with ADP-AlF4 and (dT)7ds11bp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Dai, Y.X, Chen, W.F, Teng, F.Y, Liu, N.N, Hou, X.M, Dou, S.X, Rety, S, Xi, X.G. | | Deposit date: | 2019-06-25 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and functional studies of SF1B Pif1 from Thermus oshimai reveal dimerization-induced helicase inhibition.
Nucleic Acids Res., 49, 2021
|
|
6S3E
 
 | | Crystal structure of helicase Pif1 from Thermus oshimai in apo form | | Descriptor: | PIF1 helicase | | Authors: | Dai, Y.X, Chen, W.F, Teng, F.Y, Liu, N.N, Hou, X.M, Dou, S.X, Rety, S, Xi, X.G. | | Deposit date: | 2019-06-25 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.787 Å) | | Cite: | Structural and functional studies of SF1B Pif1 from Thermus oshimai reveal dimerization-induced helicase inhibition.
Nucleic Acids Res., 49, 2021
|
|
6S3M
 
 | | Crystal structure of helicase Pif1 from Thermus oshimai in complex with ssDNA (dT)18 and ADP-AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*G)-3'), MAGNESIUM ION, ... | | Authors: | Dai, Y.X, Chen, W.F, Teng, F.Y, Liu, N.N, Hou, X.M, Dou, S.X, Rety, S, Xi, X.G. | | Deposit date: | 2019-06-25 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | Structural and functional studies of SF1B Pif1 from Thermus oshimai reveal dimerization-induced helicase inhibition.
Nucleic Acids Res., 49, 2021
|
|
6S3O
 
 | | Crystal structure of helicase Pif1 from Thermus oshimai in complex with ssDNA (dT)18 and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Dai, Y.X, Chen, W.F, Teng, F.Y, Liu, N.N, Hou, X.M, Dou, S.X, Rety, S, Xi, X.G. | | Deposit date: | 2019-06-25 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.974 Å) | | Cite: | Structural and functional studies of SF1B Pif1 from Thermus oshimai reveal dimerization-induced helicase inhibition.
Nucleic Acids Res., 49, 2021
|
|
8BNV
 
 | |
8BNX
 
 | |
5T1Q
 
 | | 2.15 Angstrom Crystal Structure of N-acetylmuramoyl-L-alanine Amidase from Staphylococcus aureus. | | Descriptor: | N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979, SODIUM ION, TRIETHYLENE GLYCOL | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bagnoli, F, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-19 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.15 Angstrom Crystal Structure of N-acetylmuramoyl-L-alanine Amidase from Staphylococcus aureus.
To Be Published
|
|
5SWU
 
 | |
5T1P
 
 | | Crystal structure of the putative periplasmic solute-binding protein from Campylobacter jejuni | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ABC transporter, ... | | Authors: | Filippova, E.V, Wawrzsak, Z, Sandoval, J, Skarina, T, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-19 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the putative periplasmic solute-binding protein from Campylobacter jejuni
To Be Published
|
|
5SWV
 
 | |
5SV5
 
 | | 1.0 Angstrom Crystal Structure of pre-Peptidase C-terminal Domain of Collagenase from Bacillus anthracis. | | Descriptor: | Microbial collagenase, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-04 | | Release date: | 2016-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | 1.0 Angstrom Crystal Structure of pre-Peptidase C-terminal Domain of Collagenase from Bacillus anthracis.
To Be Published
|
|
1U7J
 
 | | Solution structure of a diiron protein model | | Descriptor: | Four-helix bundle model, ZINC ION | | Authors: | Maglio, O, Nastri, F, Calhoun, J.R, Lahr, S, Pavone, V, DeGrado, W.F, Lombardi, A. | | Deposit date: | 2004-08-04 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Analysis and Design of Turns in alpha-Helical Hairpins
J.Mol.Biol., 346, 2005
|
|
