8C77
 
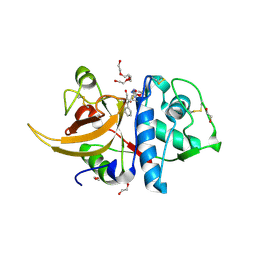 | | Human cathepsin L after reaction with the thiocarbazate inhibitor CID 16725315 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cathepsin L, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2023-01-12 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
1A6I
 
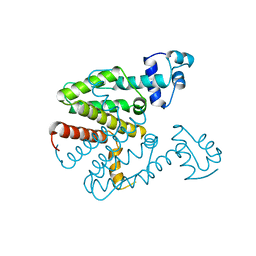 | | TET REPRESSOR, CLASS D VARIANT | | Descriptor: | TETRACYCLINE REPRESSOR PROTEIN CLASS D | | Authors: | Orth, P, Cordes, F, Schnappinger, D, Hillen, W, Saenger, W, Hinrichs, W. | | Deposit date: | 1998-02-25 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational changes of the Tet repressor induced by tetracycline trapping.
J.Mol.Biol., 279, 1998
|
|
2XBX
 
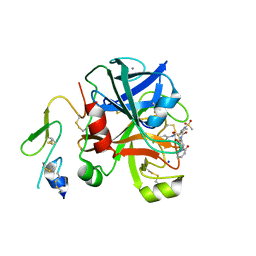 | | Factor Xa in complex with a pyrrolidine-3,4-dicarboxylic acid inhibitor | | Descriptor: | (3R,4R)-1-METHANESULFONYL-PYRROLIDINE-3,4-DICARBOXYLIC ACID 3-[(4-CHLORO-PHENYL)-AMIDE] 4-{[2-FLUORO-4-(2-OXO-2H-PYRIDIN-1-YL)-PHENYL]-AMIDE}, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Banner, D.W, Benz, J, Schlatter, D, Haap, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of a Factor Xa Inhibitor (3R,4R)-1-(2,2-Difluoro-Ethyl)-Pyrrolidine-3,4-Dicarboxylic Acid 3-[(5-Chloro-Pyridin-2-Yl)-Amide] 4-{[2-Fluoro-4-(2-Oxo-2H-Pyridin-1-Yl)-Phenyl]-Amide} as a Clinical Candidate.
Bioorg.Med.Chem., 20, 2010
|
|
2XC5
 
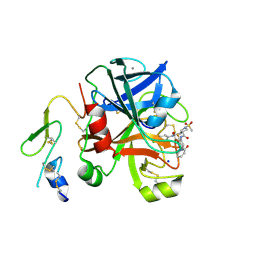 | | Factor Xa in complex with a pyrrolidine-3,4-dicarboxylic acid inhibitor | | Descriptor: | (3R,4R)-1-METHANESULFONYL-PYRROLIDINE-3,4-DICARBOXYLIC ACID 3-[(4-CHLORO-3-FLUORO-PHENYL)-AMIDE] 4-{[2-FLUORO-4-(2-OXO-2H-PYRIDIN-1-YL)-PHENYL]-AMIDE}, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Banner, D.W, Benz, J, Schlatter, D, Anselm, L, Haap, W. | | Deposit date: | 2010-04-16 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of a Factor Xa Inhibitor (3R,4R)-1-(2,2-Difluoro-Ethyl)-Pyrrolidine-3,4-Dicarboxylic Acid 3-[(5-Chloro-Pyridin-2-Yl)-Amide] 4-{[2-Fluoro-4-(2-Oxo-2H-Pyridin-1-Yl)-Phenyl]-Amide} as a Clinical Candidate.
Bioorg.Med.Chem., 20, 2010
|
|
2XBW
 
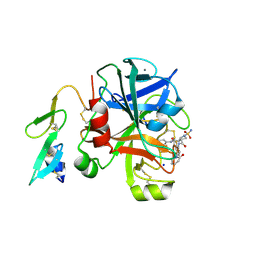 | | Factor Xa in complex with a pyrrolidine-3,4-dicarboxylic acid inhibitor | | Descriptor: | (3R,4R)-1-SULFAMOYL-PYRROLIDINE-3,4-DICARBOXYLIC ACID 3-[(4-CHLORO-PHENYL)-AMIDE] 4-{[2-FLUORO-4-(2-OXO-2H-PYRIDIN-1-YL)-PHENYL]-AMIDE}, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Banner, D.W, Benz, J, Schlatter, D, Thomi, S, Haap, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of a Factor Xa Inhibitor (3R,4R)-1-(2,2-Difluoro-Ethyl)-Pyrrolidine-3,4-Dicarboxylic Acid 3-[(5-Chloro-Pyridin-2-Yl)-Amide] 4-{[2-Fluoro-4-(2-Oxo-2H-Pyridin-1-Yl)-Phenyl]-Amide} as a Clinical Candidate.
Bioorg.Med.Chem., 20, 2010
|
|
4ZY4
 
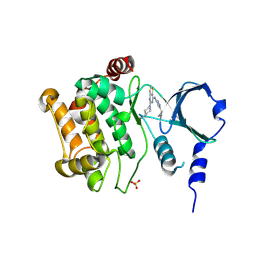 | |
2RNT
 
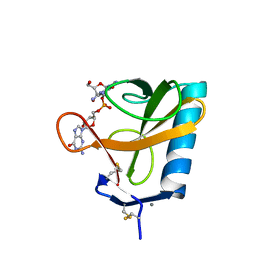 | | THREE-DIMENSIONAL STRUCTURE OF RIBONUCLEASE T1 COMPLEXED WITH GUANYLYL-2(PRIME),5(PRIME)-GUANOSINE AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, GUANYLYL-2',5'-PHOSPHOGUANOSINE, RIBONUCLEASE T1 | | Authors: | Saenger, W, Koepke, J, Maslowska, M, Heinemann, U. | | Deposit date: | 1988-07-06 | | Release date: | 1989-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of ribonuclease T1 complexed with guanylyl-2',5'-guanosine at 1.8 A resolution.
J.Mol.Biol., 206, 1989
|
|
4ZY6
 
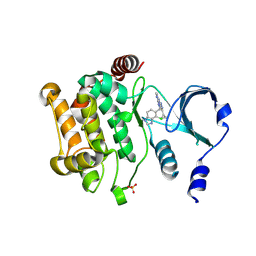 | |
2ZJK
 
 | |
2ZMM
 
 | | Crystal structure of PTP1B-inhibitor complex | | Descriptor: | 4-bromo-3-(carboxymethoxy)-5-{3-[cyclohexyl(methylcarbamoyl)amino]phenyl}thiophene-2-carboxylic acid, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Xu, W, Wu, J. | | Deposit date: | 2008-04-19 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based optimization of protein tyrosine phosphatase-1 B inhibitors: capturing interactions with arginine 24
Chemmedchem, 3, 2008
|
|
7QCG
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2-pyrrolidyl)-3,4,5-trihydroxybenzoylhydrazone | | Descriptor: | 3,4,5-tris(oxidanyl)-N-[(E)-1H-pyrrol-2-ylmethylideneamino]benzamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-23 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
1U4E
 
 | | Crystal Structure of Cytoplasmic Domains of GIRK1 channel | | Descriptor: | G protein-activated inward rectifier potassium channel 1 | | Authors: | Pegan, S, Arrabit, C, Zhou, W, Kwiatkowski, W, Slesinger, P.A, Choe, S. | | Deposit date: | 2004-07-24 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Cytoplasmic domain structures of Kir2.1 and Kir3.1 show sites for modulating gating and rectification
Nat.Neurosci., 8, 2005
|
|
1U4F
 
 | | Crystal Structure of Cytoplasmic Domains of IRK1 (Kir2.1) channel | | Descriptor: | Inward rectifier potassium channel 2 | | Authors: | Pegan, S, Arrabit, C, Zhou, W, Kwiatkowski, W, Slesinger, P.A, Choe, S. | | Deposit date: | 2004-07-24 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Cytoplasmic domain structures of Kir2.1 and Kir3.1 show sites for modulating gating and rectification
Nat.Neurosci., 8, 2005
|
|
1TPO
 
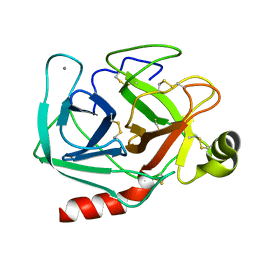 | | THE GEOMETRY OF THE REACTIVE SITE AND OF THE PEPTIDE GROUPS IN TRYPSIN, TRYPSINOGEN AND ITS COMPLEXES WITH INHIBITORS | | Descriptor: | BETA-TRYPSIN, CALCIUM ION | | Authors: | Bode, W, Walter, J, Huber, R. | | Deposit date: | 1982-09-27 | | Release date: | 1983-01-18 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Geometry of the Reactive Site and of the Peptide Groups in Trypsin, Trypsinogen and its Complexes with Inhibitors
Acta Crystallogr.,Sect.B, 39, 1983
|
|
7QKD
 
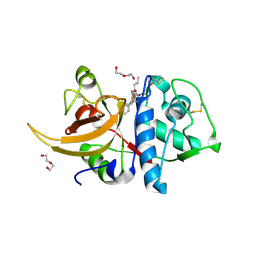 | | Crystal structure of human Cathepsin L in complex with covalently bound MG132 | | Descriptor: | ACETATE ION, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 2024
|
|
1W1I
 
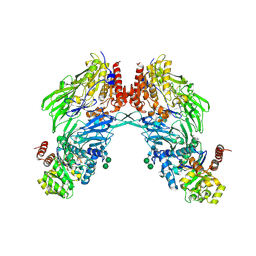 | | Crystal structure of dipeptidyl peptidase IV (DPPIV or CD26) in complex with adenosine deaminase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Weihofen, W.A, Liu, J, Reutter, W, Saenger, W, Fan, H. | | Deposit date: | 2004-06-22 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Crystal structure of CD26/dipeptidyl-peptidase IV in complex with adenosine deaminase reveals a highly amphiphilic interface.
J. Biol. Chem., 279, 2004
|
|
1IAB
 
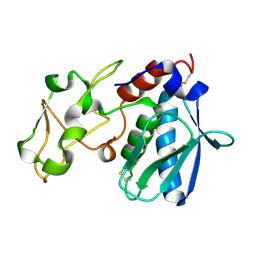 | | CRYSTAL STRUCTURES, SPECTROSCOPIC FEATURES, AND CATALYTIC PROPERTIES OF COBALT(II), COPPER(II), NICKEL(II), AND MERCURY(II) DERIVATIVES OF THE ZINC ENDOPEPTIDASE ASTACIN. A CORRELATION OF STRUCTURE AND PROTEOLYTIC ACTIVITY | | Descriptor: | ASTACIN, COBALT (II) ION | | Authors: | Gomis-Rueth, F.-X, Stoecker, W, Bode, W. | | Deposit date: | 1994-05-09 | | Release date: | 1994-08-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structures, spectroscopic features, and catalytic properties of cobalt(II), copper(II), nickel(II), and mercury(II) derivatives of the zinc endopeptidase astacin. A correlation of structure and proteolytic activity.
J.Biol.Chem., 269, 1994
|
|
1IAE
 
 | | CRYSTAL STRUCTURES, SPECTROSCOPIC FEATURES, AND CATALYTIC PROPERTIES OF COBALT(II), COPPER(II), NICKEL(II), AND MERCURY(II) DERIVATIVES OF THE ZINC ENDOPEPTIDASE ASTACIN. A CORRELATION OF STRUCTURE AND PROTEOLYTIC ACTIVITY | | Descriptor: | ASTACIN, NICKEL (II) ION | | Authors: | Grams, F, Stoecker, W, Bode, W. | | Deposit date: | 1994-05-09 | | Release date: | 1994-08-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures, spectroscopic features, and catalytic properties of cobalt(II), copper(II), nickel(II), and mercury(II) derivatives of the zinc endopeptidase astacin. A correlation of structure and proteolytic activity.
J.Biol.Chem., 269, 1994
|
|
1IAD
 
 | | REFINED 1.8 ANGSTROMS X-RAY CRYSTAL STRUCTURE OF ASTACIN, A ZINC-ENDOPEPTIDASE FROM THE CRAYFISH ASTACUS ASTACUS L. STRUCTURE DETERMINATION, REFINEMENT, MOLECULAR STRUCTURE AND COMPARISON TO THERMOLYSIN | | Descriptor: | ASTACIN | | Authors: | Gomis-Rueth, F.-X, Stoecker, W, Bode, W. | | Deposit date: | 1994-05-09 | | Release date: | 1994-08-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined 1.8 A X-ray crystal structure of astacin, a zinc-endopeptidase from the crayfish Astacus astacus L. Structure determination, refinement, molecular structure and comparison with thermolysin.
J.Mol.Biol., 229, 1993
|
|
6O6W
 
 | | Solution structure of human myeloid-derived growth factor | | Descriptor: | Myeloid-derived growth factor | | Authors: | Bortnov, V, Tonelli, M, Lee, W, Markley, J.L, Mosher, D.F. | | Deposit date: | 2019-03-07 | | Release date: | 2019-11-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human myeloid-derived growth factor suggests a conserved function in the endoplasmic reticulum.
Nat Commun, 10, 2019
|
|
1IAA
 
 | | CRYSTAL STRUCTURES, SPECTROSCOPIC FEATURES, AND CATALYTIC PROPERTIES OF COBALT(II), COPPER(II), NICKEL(II), AND MERCURY(II) DERIVATIVES OF THE ZINC ENDOPEPTIDASE ASTACIN. A CORRELATION OF STRUCTURE AND PROTEOLYTIC ACTIVITY | | Descriptor: | ASTACIN, COPPER (II) ION | | Authors: | Gomis-Rueth, F.-X, Stoecker, W, Bode, W. | | Deposit date: | 1994-05-09 | | Release date: | 1994-08-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures, spectroscopic features, and catalytic properties of cobalt(II), copper(II), nickel(II), and mercury(II) derivatives of the zinc endopeptidase astacin. A correlation of structure and proteolytic activity.
J.Biol.Chem., 269, 1994
|
|
5U5H
 
 | | Crystal structure of EED in complex with 6-(2-fluoro-5-methoxybenzyl)-1-isopropyl-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-3-amine 6-(2-fluoro-5-methoxybenzyl)-1-isopropyl-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-3-amine | | Descriptor: | (6S)-6-[(2-fluoro-5-methoxyphenyl)methyl]-1-(propan-2-yl)-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-3-amine, GLYCEROL, Polycomb protein EED | | Authors: | Bussiere, D, Shu, W. | | Deposit date: | 2016-12-06 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Design of EED Binders Allosterically Inhibiting the Epigenetic Polycomb Repressive Complex 2 (PRC2) Methyltransferase.
J. Med. Chem., 60, 2017
|
|
6PMJ
 
 | | Sigm28-transcription initiation complex with specific promoter at the state 2 | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Shi, W. | | Deposit date: | 2019-07-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structural basis of bacterial sigma28-mediated transcription reveals roles of the RNA polymerase zinc-binding domain.
Embo J., 39, 2020
|
|
7NFV
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Srinivasan, V, Gunther, S, Reinke, P, Werner, N, Falke, S, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Sprenger, J, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Koua, F, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Ewert, W, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Ehrt, C, Rarey, M, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Hinrichs, W, Meents, A, Betzel, C. | | Deposit date: | 2021-02-07 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
3J7W
 
 | | Capsid Expansion Mechanism Of Bacteriophage T7 Revealed By Multi-State Atomic Models Derived From Cryo-EM Reconstructions | | Descriptor: | Major capsid protein 10A | | Authors: | Guo, F, Liu, Z, Fang, P.A, Zhang, Q, Wright, E.T, Wu, W, Zhang, C, Vago, F, Ren, Y, Jakata, J, Chiu, W, Serwer, P, Jiang, W. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Capsid expansion mechanism of bacteriophage T7 revealed by multistate atomic models derived from cryo-EM reconstructions.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
