5MJ7
 
 | | Structure of the C. elegans nucleoside hydrolase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Uncharacterized protein | | Authors: | Versees, W, Singh, R.K. | | Deposit date: | 2016-11-30 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and biochemical characterization of the nucleoside hydrolase from C. elegans reveals the role of two active site cysteine residues in catalysis.
Protein Sci., 26, 2017
|
|
5MBR
 
 | |
5MCR
 
 | | Quadruplex with flipped tetrad formed by an artificial sequence | | Descriptor: | Artificial quadruplex with propeller, diagonal and lateral loop | | Authors: | Dickerhoff, J, Haase, L, Langel, W, Weisz, K. | | Deposit date: | 2016-11-10 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Tracing Effects of Fluorine Substitutions on G-Quadruplex Conformational Changes.
ACS Chem. Biol., 12, 2017
|
|
5MPE
 
 | | 26S proteasome in presence of ATP (s2) | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit RPN1, 26S proteasome regulatory subunit RPN10, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5M82
 
 | | Three-dimensional structure of the photoproduct state of GAF3 from Slr1393 of Synechocystis sp. PCC6803 | | Descriptor: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, PHYCOCYANOBILIN, SODIUM ION, ... | | Authors: | Xu, X.-L, Zhao, K.-H, Gaertner, W, Hoeppner, A. | | Deposit date: | 2016-10-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural elements regulating the photochromicity in a cyanobacteriochrome
Proc.Natl.Acad.Sci.USA, 2020
|
|
6CIL
 
 | | PRE-REACTION COMPLEX, RAG1(E962Q)/2-INTACT/INTACT 12/23RSS COMPLEX IN MN2+ | | Descriptor: | High mobility group protein B1, Intact 12RSS substrate forward strand, Intact 12RSS substrate reverse strand, ... | | Authors: | Chuenchor, W, Chen, X, Kim, M.S, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
5MPA
 
 | | 26S proteasome in presence of ATP (s2) | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MPC
 
 | | 26S proteasome in presence of BeFx (s4) | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MFP
 
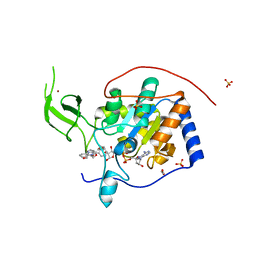 | | Human Sirt6 in complex with activator UBCS58 | | Descriptor: | 1,2-ETHANEDIOL, 4-pyridin-3-ylpyrrolo[1,2-a]quinoxaline, NAD-dependent protein deacetylase sirtuin-6, ... | | Authors: | Steegborn, C, You, W, Kambach, C. | | Deposit date: | 2016-11-18 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Basis of Sirtuin 6 Activation by Synthetic Small Molecules.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MG5
 
 | | A multi-component acyltransferase PhlABC from Pseudomonas protegens soaked with the monoacetylphloroglucinol (MAPG) | | Descriptor: | 2,4-diacetylphloroglucinol biosynthesis protein, 2,4-diacetylphloroglucinol biosynthesis protein PhlC, Hydroxymethylglutaryl-CoA synthase, ... | | Authors: | Pavkov-Keller, T, Schmidt, N.G, Kroutil, W, Gruber, K. | | Deposit date: | 2016-11-20 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.44 Å) | | Cite: | Structure and Catalytic Mechanism of a Bacterial Friedel-Crafts Acylase.
Chembiochem, 20, 2019
|
|
6CIJ
 
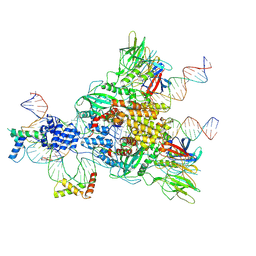 | | Cryo-EM structure of mouse RAG1/2 HFC complex containing partial HMGB1 linker(3.9 A) | | Descriptor: | CALCIUM ION, DNA (30-MER), DNA (41-MER), ... | | Authors: | Chen, X, Kim, M, Chuenchor, W, Cui, Y, Zhang, X, Zhou, Z.H, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
5MTR
 
 | | Crystal structure of M. tuberculosis InhA inhibited by PT512 | | Descriptor: | 2-[4-[(4-cyclopentyl-1,2,3-triazol-1-yl)methyl]-2-oxidanyl-phenoxy]benzenecarbonitrile, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
4V0I
 
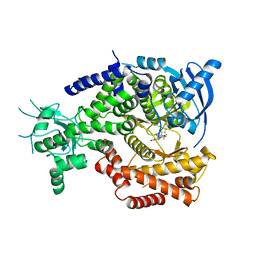 | | Water Network Determines Selectivity for a Series of Pyrimidone Indoline Amide PI3KBeta Inhibitors over PI3K-Delta | | Descriptor: | 2-[2-(2-METHYL-2,3-DIHYDRO-INDOL-1-YL)-2-OXO-ETHYL]-6-MORPHOLIN-4-YL-3H-PYRIMIDIN-4-ONE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Robinson, D, Bertrand, T, Carry, J.C, Halley, F, Karlsson, A, Mathieu, M, Minoux, H, Perrin, M.A, Robert, B, Schio, L, Sherman, W. | | Deposit date: | 2014-09-16 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Differential Water Thermodynamics Determine Pi3K-Beta/Delta Selectivity for Solvent-Exposed Ligand Modifications.
J.Chem.Inf.Model., 56, 2016
|
|
4W5U
 
 | |
4V1N
 
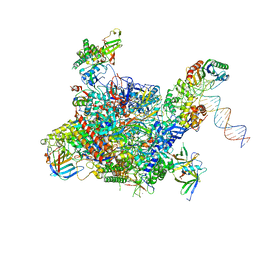 | | Architecture of the RNA polymerase II-Mediator core transcription initiation complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Lariviere, L, Wenzeck, L, Hemann, M, Tegunov, D, Petrotchenko, E.V, Borchers, C.H, Baumeister, W, Herzog, F, Villa, E, Cramer, P. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Architecture of the RNA Polymerase II-Mediator Core Initiation Complex.
Nature, 518, 2015
|
|
4UXV
 
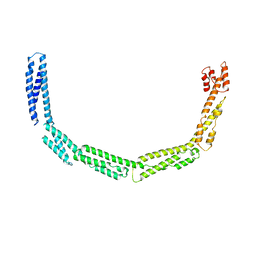 | | Cytoplasmic domain of bacterial cell division protein EzrA | | Descriptor: | SEPTATION RING FORMATION REGULATOR EZRA | | Authors: | Cleverley, R.M, Barrett, J.R, Basle, A, Khai-Bui, N, Hewitt, L, Solovyova, A, Xu, Z, Daniela, R.A, Dixon, N.E, Harry, E.J, Oakley, A.J, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-08-27 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.961 Å) | | Cite: | Structure and Function of a Spectrin-Like Regulator of Bacterial Cytokinesis.
Nat.Commun., 5, 2014
|
|
4WB0
 
 | |
4UV3
 
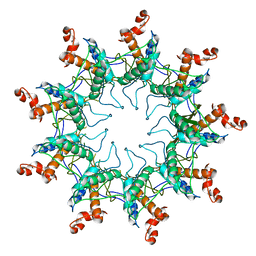 | | Structure of the curli transport lipoprotein CsgG in its membrane- bound conformation | | Descriptor: | CURLI PRODUCTION ASSEMBLY/TRANSPORT COMPONENT CSGG | | Authors: | Goyal, P, Krasteva, P.V, Gerven, N.V, Gubellini, F, Broeck, I.V.D, Troupiotis-Tsailaki, A, Jonckheere, W, Pehau-Arnaudet, G, Pinkner, J.S, Chapman, M.R, Hultgren, S.J, Howorka, S, Fronzes, R, Remaut, H. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural and Mechanistic Insights Into the Bacterial Amyloid Secretion Channel Csgg.
Nature, 516, 2014
|
|
4V1O
 
 | | Architecture of the RNA polymerase II-Mediator core transcription initiation complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Lariviere, L, Wenzeck, L, Hemann, M, Tegunov, D, Petrotchenko, E.V, Borchers, C.H, Baumeister, W, Herzog, F, Villa, E, Cramer, P. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Architecture of the RNA Polymerase II-Mediator Core Initiation Complex.
Nature, 518, 2015
|
|
4W7H
 
 | | Crystal Structure of DEH Reductase A1-R Mutant | | Descriptor: | Carbonyl reductase | | Authors: | Takase, R, Mikami, B, Kawai, S, Murata, K, Hashimoto, W. | | Deposit date: | 2014-08-22 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structure-based Conversion of the Coenzyme Requirement of a Short-chain Dehydrogenase/Reductase Involved in Bacterial Alginate Metabolism.
J.Biol.Chem., 289, 2014
|
|
4URD
 
 | | Cryo-EM map of Trigger Factor bound to a translating ribosome | | Descriptor: | TRIGGER FACTOR | | Authors: | Deeng, J, Chan, K.Y, van der Sluis, E, Bischoff, L, Berninghausen, O, Han, W, Gumbart, J, Schulten, K, Beatrix, B, Beckmann, R. | | Deposit date: | 2014-06-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Dynamic Behavior of Trigger Factor on the Ribosome.
J.Mol.Biol., 428, 2016
|
|
4V4N
 
 | | Structure of the Methanococcus jannaschii ribosome-SecYEBeta channel complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein L7AE, ... | | Authors: | Menetret, J.F, Park, E, Gumbart, J.C, Ludtke, S.J, Li, W, Whynot, A, Rapoport, T.A, Akey, C.W. | | Deposit date: | 2013-06-17 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure of the SecY channel during initiation of protein translocation.
Nature, 506, 2013
|
|
4V6E
 
 | | Crystal structure of the E. coli 70S ribosome in an intermediate state of ratcheting | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, W, Dunkle, J.A, Cate, J.H.D. | | Deposit date: | 2009-06-28 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.712 Å) | | Cite: | Structures of the ribosome in intermediate States of ratcheting.
Science, 325, 2009
|
|
4V94
 
 | | Molecular architecture of the eukaryotic chaperonin TRiC/CCT derived by a combination of chemical crosslinking and mass-spectrometry, XL-MS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Leitner, A, Joachimiak, L.A, Bracher, A, Walzthoeni, T, Chen, B, Monkemeyer, L, Pechmann, S, Holmes, S, Cong, Y, Ma, B, Ludtke, S, Chiu, W, Hartl, F.U, Aebersold, R, Frydman, J. | | Deposit date: | 2012-01-11 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Molecular Architecture of the Eukaryotic Chaperonin TRiC/CCT.
Structure, 20, 2012
|
|
4V50
 
 | | Crystal Structure of Ribosome with messenger RNA and the Anticodon stem-loop of P-site tRNA. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Berk, V, Zhang, W, Pai, R.D, Cate, J.H.D. | | Deposit date: | 2006-08-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural basis for mRNA and tRNA positioning on the ribosome.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
