7ESA
 
 | | the complex structure of flavin transferase FmnB complexed with FAD | | Descriptor: | FAD:protein FMN transferase, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Zheng, Y.H, Cheng, W. | | Deposit date: | 2021-05-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
1FK5
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH OLEIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN, OLEIC ACID | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FKG
 
 | | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS, AND THE X-RAY CRYSTAL STRUCTURES OF THEIR COMPLEXES WITH FKBP12 | | Descriptor: | 1,3-DIPHENYL-1-PROPYL-1-(3,3-DIMETHYL-1,2-DIOXYPENTYL)-2-PIPERIDINE CARBOXYLATE, FK506 BINDING PROTEIN | | Authors: | Holt, D.A, Luengo, J.I, Yamashita, D.S, Oh, H.-J, Konialian, A.L, Yen, H.-K, Rozamus, L.W, Brandt, M, Bossard, M.J, Levy, M.A, Eggleston, D.S, Stout, T.J, Liang, J, Schultz, L.W, Clardy, J. | | Deposit date: | 1993-08-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS AND THE X-RAY CRYSTAL-STRUCTURES OF THEIR COMPLEXES WITH FKBP12.
J.Am.Chem.Soc., 115, 1993
|
|
3M9W
 
 | |
2MRT
 
 | | CONFORMATION OF CD-7 METALLOTHIONEIN-2 FROM RAT LIVER IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2 | | Authors: | Braun, W, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformation of [Cd7]-metallothionein-2 from rat liver in aqueous solution determined by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 203, 1988
|
|
7ESB
 
 | | FmnB complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FAD:protein FMN transferase, MAGNESIUM ION | | Authors: | Zheng, Y.H, Cheng, W. | | Deposit date: | 2021-05-09 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
6HQO
 
 | | Crystal structure of GcoA F169S bound to guaiacol | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3LZI
 
 | | RB69 DNA Polymerase (Y567A) ternary complex with dATP Opposite 7,8-dihydro-8-oxoguanine | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | Authors: | Wang, M, Beckman, J, Blaha, G, Wang, J, Konigsberg, W.H. | | Deposit date: | 2010-03-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substitution of Ala for Tyr567 in RB69 DNA polymerase allows dAMP to be inserted opposite 7,8-dihydro-8-oxoguanine .
Biochemistry, 49, 2010
|
|
4ARA
 
 | | Mus musculus Acetylcholinesterase in complex with (R)-C5685 at 2.5 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(DIMETHYLAMINO)-N-{[(2R)-1-ETHYLPYRROLIDIN-2-YL]METHYL}-2-METHOXY-5-NITROBENZAMIDE, ... | | Authors: | Berg, L, Niemiec, M.S, Qian, W, Andersson, C.D, WittungStafshede, P, Ekstrom, F, Linusson, A. | | Deposit date: | 2012-04-23 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Similar But Different: Thermodynamic and Structural Characterization of a Pair of Enantiomers Binding to Acetylcholinesterase.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4AIT
 
 | |
8RM7
 
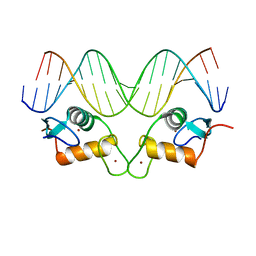 | | Crystal Structure of Human Androgen Receptor DNA Binding Domain Bound to its Response Element: MMTV-177 GRE/ARE | | Descriptor: | Isoform 2 of Androgen receptor, MMTV-177 GRE/ARE Chain C, MMTV-177 GRE/ARE, ... | | Authors: | Lee, X.Y, Helsen, C, Van Eynde, W, Voet, A, Claessens, F. | | Deposit date: | 2024-01-05 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural mechanism underlying variations in DNA binding by the androgen receptor.
J.Steroid Biochem.Mol.Biol., 241, 2024
|
|
8RM6
 
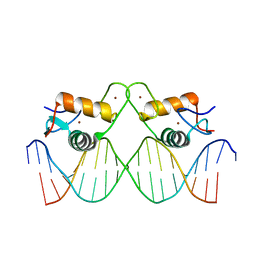 | | Crystal Structure of Human Androgen Receptor DNA Binding Domain Bound to its Response Element: C3(1)ARE | | Descriptor: | C3(1)ARE_Chain C, C3(1)ARE_Chain D, Isoform 2 of Androgen receptor, ... | | Authors: | Lee, X.Y, Helsen, C, Van Eynde, W, Voet, A, Claessens, F. | | Deposit date: | 2024-01-05 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural mechanism underlying variations in DNA binding by the androgen receptor.
J.Steroid Biochem.Mol.Biol., 241, 2024
|
|
1FRP
 
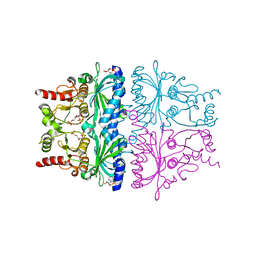 | | CRYSTAL STRUCTURE OF FRUCTOSE-1,6-BISPHOSPHATASE COMPLEXED WITH FRUCTOSE-2,6-BISPHOSPHATE, AMP AND ZN2+ AT 2.0 ANGSTROMS RESOLUTION. ASPECTS OF SYNERGISM BETWEEN INHIBITORS | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE, ... | | Authors: | Xue, Y, Huang, S, Liang, J.-Y, Zhang, Y, Lipscomb, W.N. | | Deposit date: | 1994-08-26 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of fructose-1,6-bisphosphatase complexed with fructose 2,6-bisphosphate, AMP, and Zn2+ at 2.0-A resolution: aspects of synergism between inhibitors.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
8R35
 
 | |
1FPE
 
 | | STRUCTURAL ASPECTS OF THE ALLOSTERIC INHIBITION OF FRUCTOSE-1,6-BISPHOSPHATASE BY AMP: THE BINDING OF BOTH THE SUBSTRATE ANALOGUE 2,5-ANHYDRO-D-GLUCITOL-1,6-BISPHOSPHATE AND CATALYTIC METAL IONS MONITORED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE, ... | | Authors: | Villeret, V, Huang, S, Zhang, Y, Lipscomb, W.N. | | Deposit date: | 1994-12-15 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural aspects of the allosteric inhibition of fructose-1,6-bisphosphatase by AMP: the binding of both the substrate analogue 2,5-anhydro-D-glucitol 1,6-bisphosphate and catalytic metal ions monitored by X-ray crystallography.
Biochemistry, 34, 1995
|
|
2W63
 
 | |
8R4C
 
 | |
2WDZ
 
 | | Crystal structure of the short chain dehydrogenase Galactitol- Dehydrogenase (GatDH) of Rhodobacter sphaeroides in complex with NAD+ and 1,2-Pentandiol | | Descriptor: | (2S)-pentane-1,2-diol, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Carius, Y, Christian, H, Faust, A, Kornberger, P, Kohring, G.W, Giffhorn, F, Scheidig, A.J. | | Deposit date: | 2009-03-27 | | Release date: | 2010-03-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insight Into Substrate Differentiation of the Sugar-Metabolizing Enzyme Galactitol Dehydrogenase from Rhodobacter Sphaeroides D.
J.Biol.Chem., 285, 2010
|
|
7K1D
 
 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound BDM_77291 | | Descriptor: | (3R)-3-{4-[(3R)-4-(3,4-difluorobenzene-1-carbonyl)morpholin-3-yl]-1H-1,2,3-triazol-1-yl}-N-hydroxy-4-(naphthalen-2-yl)butanamide, 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Liang, W.G, Deprez, R, Bosc, D, Tang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 2
To Be Published
|
|
1PVD
 
 | |
7K1F
 
 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound BDM_88558 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 3,4-difluoro-N-({1-[(2R)-4-(hydroxyamino)-4-oxo-1-(quinolin-7-yl)butan-2-yl]-1H-1,2,3-triazol-4-yl}methyl)benzamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Liang, W.G, Deprez, R, Bosc, D, Tang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 4
To Be Published
|
|
2W61
 
 | |
8R34
 
 | | CryoEM structure of the symmetric Pho90 dimer from yeast with substrates. | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, Low-affinity phosphate transporter PHO90, PHOSPHATE ION, ... | | Authors: | Schneider, S, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2023-11-08 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Complementary structures of the yeast phosphate transporter Pho90 provide insights into its transport mechanism.
Structure, 32, 2024
|
|
7WOR
 
 | | The state 2 of Omicron Spike with bispecific antibody FD01 | | Descriptor: | 16L9 Fv, 2-acetamido-2-deoxy-beta-D-glucopyranose, GW01 Fv, ... | | Authors: | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
8PZQ
 
 | | Model for focused reconstruction of influenza A RNP-like particle | | Descriptor: | Nucleoprotein, RNA (5'P-(UC)6-FAM3') | | Authors: | Chenavier, F, Estrozi, L.F, Zarkadas, E, Ruigrok, R.W.H, Schoehn, G, Ballandras-Colas, A, Crepin, T. | | Deposit date: | 2023-07-27 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Cryo-EM structure of influenza helical nucleocapsid reveals NP-NP and NP-RNA interactions as a model for the genome encapsidation.
Sci Adv, 9, 2023
|
|
