7S6Y
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI32 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-[(cyclopropylmethyl)amino]-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S75
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI42 | | Descriptor: | (1R,2S,5S)-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(3-methylbutanoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S74
 
 | |
7S6Z
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI33 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-(ethylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S73
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI37 | | Descriptor: | (6S)-5-{(2S)-2-[(tert-butylcarbamoyl)amino]-3,3-dimethylbutanoyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-5-azaspiro[2.4]heptane-6-carboxamide (non-preferred name), 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
6YSS
 
 | | Structure of the P+9 ArfB-ribosome complex in the post-hydrolysis state | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
6YST
 
 | | Structure of the P+9 ArfB-ribosome complex with P/E hybrid tRNA in the post-hydrolysis state | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
6YSU
 
 | | Structure of the P+0 ArfB-ribosome complex in the post-hydrolysis state | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
6YSR
 
 | | Structure of the P+9 stalled ribosome complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
6LHO
 
 | | The cryo-EM structure of coxsackievirus A16 empty particle in complex with Fab 18A7 | | Descriptor: | VP1 protein, VP2 protein, VP3 protein | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
4G9B
 
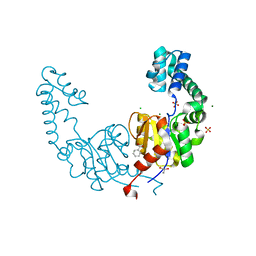 | | Crystal structure of beta-phosphoglucomutase homolog from escherichia coli, target efi-501172, with bound mg, open lid | | Descriptor: | Beta-phosphoglucomutase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of beta-phosphoglucomutase homolog from escherichia coli, target efi-501172, with bound mg, open lid
To be Published
|
|
4GE1
 
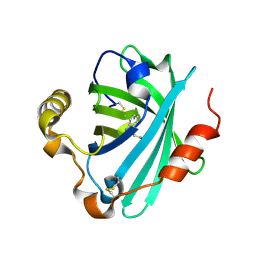 | | Structure of the tryptamine complex of the amine binding protein of Rhodnius prolixus | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANAMINE, Biogenic amine-binding protein, GLYCEROL | | Authors: | Andersen, J.F, Chang, B.W, Xu, X, Mans, B.J, Ribeiro, J.M. | | Deposit date: | 2012-08-01 | | Release date: | 2013-01-02 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and ligand-binding properties of the biogenic amine-binding protein from the saliva of a blood-feeding insect vector of Trypanosoma cruzi.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6L08
 
 | |
3CZF
 
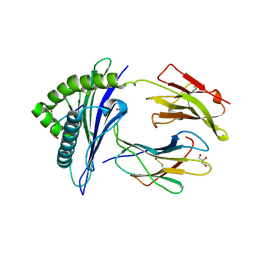 | | Crystal structure of HLA-B*2709 complexed with the glucagon receptor (GR) peptide (residues 412-420) | | Descriptor: | BETA-2-MICROGLOBULIN, GLUCAGON RECEPTOR PEPTIDE, GLYCEROL, ... | | Authors: | Loll, B, Fiorillo, M.T, Rueckert, C, Saenger, W, Sorrentino, R, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2008-04-29 | | Release date: | 2009-04-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Conformational Plasticity of HLA-B27 Molecules Correlates Inversely With Efficiency of Negative T Cell Selection.
Front Immunol, 11, 2020
|
|
4G39
 
 | |
4FYE
 
 | | Crystal structure of a Legionella phosphoinositide phosphatase, SidF | | Descriptor: | PHOSPHATE ION, SidF, inhibitor of growth family, ... | | Authors: | Hsu, F.S, Zhu, W, Brennan, L, Tao, L, Luo, Z.Q, Mao, Y. | | Deposit date: | 2012-07-04 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structural basis for substrate recognition by a unique Legionella phosphoinositide phosphatase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GDB
 
 | | SHV-1 in complex with 4H-pyrazolo[1,5-c][1,3]thiazole containing penem inhibitor | | Descriptor: | (7R)-6-formyl-7-(4H-pyrazolo[1,5-c][1,3]thiazol-2-yl)-4,7-dihydro-1,4-thiazepine-3-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase SHV-1, ... | | Authors: | Ke, W, van den Akker, F. | | Deposit date: | 2012-07-31 | | Release date: | 2013-07-31 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structures of SHV-1 beta-lactamase with penem and penam sulfone inhibitors that form cyclic intermediates stabilized by carbonyl conjugation
Plos One, 7, 2012
|
|
4G1T
 
 | | Crystal structure of interferon-stimulated gene 54 | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 2 | | Authors: | Yang, Z, Liang, H, Zhou, Q, Li, Y, Chen, H, Ye, W, Chen, D, Fleming, J, Shu, H, Liu, Y. | | Deposit date: | 2012-07-11 | | Release date: | 2012-08-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ISG54 reveals a novel RNA binding structure and potential functional mechanisms.
Cell Res., 22, 2012
|
|
4GET
 
 | | Crystal structure of biogenic amine binding protein from Rhodnius prolixus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Biogenic amine-binding protein | | Authors: | Andersen, J.F, Chang, B.W, Xu, X, Mans, B.J, Ribeiro, J.M. | | Deposit date: | 2012-08-02 | | Release date: | 2013-01-02 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure and ligand-binding properties of the biogenic amine-binding protein from the saliva of a blood-feeding insect vector of Trypanosoma cruzi.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3CL4
 
 | | Crystal structure of bovine coronavirus hemagglutinin-esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-esterase, ... | | Authors: | Zeng, Q.H, Langereis, M.A, van Vliet, A.L.W, Huizinga, E.G, de Groot, R.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6LIX
 
 | | CRL Protein of Arabidopsis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chromophore lyase CRL, chloroplastic | | Authors: | Wang, F.F, Guan, K.L, Sun, P.K, Xing, W.M. | | Deposit date: | 2019-12-13 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | The Arabidopsis CRUMPLED LEAF protein, a homolog of the cyanobacterial bilin lyase, retains the bilin-binding pocket for a yet unknown function.
Plant J., 104, 2020
|
|
4G4X
 
 | | Crystal structure of peptidoglycan-associated lipoprotein from Acinetobacter baumannii | | Descriptor: | D-ALANINE, GLYCEROL, Peptidoglycan-associated lipoprotein, ... | | Authors: | Lee, W.C, Song, J.H, Park, J.S, Kim, H.Y. | | Deposit date: | 2012-07-16 | | Release date: | 2013-07-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enantiomer-dependent amino acid binding affinity of OmpA-like domains from Acinetobacter baumannii peptidoglycan-associated lipoprotein and OmpA
To be Published
|
|
4GME
 
 | | Crystal structure of mannonate dehydratase (target EFI-502209) from caulobacter crescentus cb15 complexed with magnesium and d-mannonate | | Descriptor: | CARBONATE ION, CHLORIDE ION, D-MANNONIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-15 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Mannonate Dehydratase from Caulobacter Crescentus Cb15
To be Published
|
|
6LD9
 
 | |
4GBR
 
 | |
