4B4U
 
 | | Crystal structure of Acinetobacter baumannii N5, N10- methylenetetrahydrofolate dehydrogenase-cyclohydrolase (FolD) complexed with NADP cofactor | | Descriptor: | 1,2-ETHANEDIOL, BIFUNCTIONAL PROTEIN FOLD, CHLORIDE ION, ... | | Authors: | Eadsforth, T.C, Maluf, F.V, Hunter, W.N. | | Deposit date: | 2012-08-01 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Acinetobacter Baumannii Fold Ligand Complexes; Potent Inhibitors of Folate Metabolism and a Re-Evaluation of the Ly374571 Structure.
FEBS J., 279, 2012
|
|
4B8P
 
 | | rImp_alpha_A89NLS | | Descriptor: | A89NLS, IMPORTIN SUBUNIT ALPHA-1A | | Authors: | Chang, C.-W, Counago, R.L.M, Williams, S.J, Boden, M, Kobe, B. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Rice Importin-Alpha and Structural Basis of its Interaction with Plant-Specific Nuclear Localization Signals.
Plant Cell, 24, 2012
|
|
8GZE
 
 | | Crystal Structure of human METTL9-SAH-SLC39A7 peptide complex | | Descriptor: | Protein-L-histidine N-pros-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, Zinc transporter SLC39A7 | | Authors: | Zhao, W.T, Li, H.T. | | Deposit date: | 2022-09-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular basis for protein histidine N1-specific methylation of the "His-x-His" motifs by METTL9.
Cell Insight, 2, 2023
|
|
4BAU
 
 | | Structure of a putative epoxide hydrolase t131d mutant from Pseudomonas aeruginosa, with bound MFA | | Descriptor: | CHLORIDE ION, PROBABLE EPOXIDE HYDROLASE, SULFATE ION, ... | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2012-09-16 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of a Putative Epoxide Hydrolase T131D Mutant from Pseudomonas Aeruginosa, with Bound Mfa
To be Published
|
|
4BJU
 
 | | Genetic and structural validation of Aspergillus fumigatus N- acetylphosphoglucosamine mutase as an antifungal target | | Descriptor: | MAGNESIUM ION, N-ACETYLGLUCOSAMINE-PHOSPHATE MUTASE | | Authors: | Fang, W, Raimi, O.G, Hurtado Guerrero, R, van Aalten, D.M.F. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Genetic and Structural Validation of Aspergillus Fumigatus N-Acetylphosphoglucosamine Mutase as an Antifungal Target.
Biosci.Rep, 33, 2013
|
|
4BGC
 
 | |
4BJO
 
 | | Nitrate in the active site of PTP1b is a putative mimetic of the transition state | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Kenny, P.W, Newman, J, Peat, T.S. | | Deposit date: | 2013-04-19 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Nitrate in the Active Site of Protein Tyrosine Phosphatase 1B is a Putative Mimetic of the Transition State.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4BK3
 
 | | Crystal structure of 3-hydroxybenzoate 6-hydroxylase uncovers lipid- assisted flavoprotein strategy for regioselective aromatic hydroxylation: Y105F mutant | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATIDYLGLYCEROL-PHOSPHOGLYCEROL, ... | | Authors: | Orru, R, Montersino, S, Barendregt, A, Westphal, A.H, van Duijn, E, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2013-04-21 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of 3-Hydroxybenzoate 6-Hydroxylase Uncovers Lipid-Assisted Flavoprotein Strategy for Regioselective Aromatic Hydroxylation
J.Biol.Chem., 288, 2013
|
|
4UFW
 
 | | Plasmodium vivax N-myristoyltransferase in complex with a pyridyl inhibitor (compound 22) | | Descriptor: | 2-oxopentadecyl-CoA, 4-chloranyl-N-[2-(3-methoxyphenyl)ethanimidoyl]-2-piperidin-4-yloxy-benzamide, CHLORIDE ION, ... | | Authors: | Yu, Z, Brannigan, J.A, Rangachari, K, Heal, W.P, Wilkinson, A.J, Holder, A.A, Tate, E.W, Leatherbarrow, R.J. | | Deposit date: | 2015-03-19 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Pyridyl-Based Inhibitors of Plasmodium Falciparum N-Myristoyltransferase
Medchemcomm, 6, 2015
|
|
8HE5
 
 | | RNA polymerase II elongation complex bound with Rad26 and Elf1, stalled at SHL(-3.5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA repair protein, DNA-directed RNA polymerase subunit, ... | | Authors: | Osumi, K, Kujirai, T, Ehara, H, Kinoshita, C, Saotome, M, Kagawa, W, Sekine, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-11-07 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.95 Å) | | Cite: | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
8HL7
 
 | |
4UPF
 
 | | Assembly principles of the unique cage formed by the ATPase RavA hexamer and the lysine decarboxylase LdcI decamer | | Descriptor: | ATPASE RAVA, LYSINE DECARBOXYLASE, INDUCIBLE | | Authors: | Malet, H, Liu, K, El Bakkouri, M, Chan, S.W.S, Effantin, G, Bacia, M, Houry, W.A, Gutsche, I. | | Deposit date: | 2014-06-16 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Assembly Principles of a Unique Cage Formed by Hexameric and Decameric E. Coli Proteins.
Elife, 3, 2014
|
|
8HNJ
 
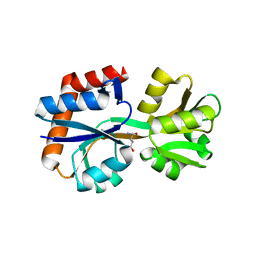 | | Domain-stabilized glutamine-binding protein | | Descriptor: | GLUTAMINE, Glutamine ABC transporter, periplasmic glutamine-binding protein GlnH | | Authors: | Choi, S.H, Park, J.H, Seo, M.H, Park, K.W, Lee, W.K. | | Deposit date: | 2022-12-08 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Domain-wise dissection of thermal stability enhancement in multidomain proteins.
Int.J.Biol.Macromol., 237, 2023
|
|
7CHY
 
 | |
7CHZ
 
 | |
4UJ7
 
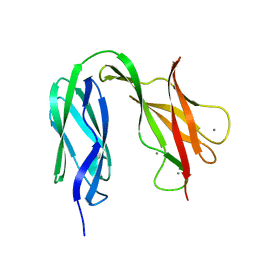 | | Structure of the S-layer protein SbsC, domains 5-6 | | Descriptor: | CALCIUM ION, SURFACE LAYER PROTEIN | | Authors: | Dordic, A, Pavkov-Keller, T, Eder, M, Egelseer, E.M, Davis, K, Mills, D, Sleytr, U.B, Kuehlbrandt, W, Vonck, J, Keller, W. | | Deposit date: | 2015-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure of the S-Layer Protein Sbsc, Domains 5-6
To be Published
|
|
4UIC
 
 | |
8TMS
 
 | | Crystal structure of bacterial pectin methylesterase PmeC2 from rumen Butyrivibrio | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Pectinesterase | | Authors: | Carbone, V, Reilly, K, Sang, C, Schofield, L, Ronimus, R, Kelly, W.J, Attwood, G.T, Palevich, N. | | Deposit date: | 2023-07-30 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Bacterial Pectin Methylesterases Pme8A and PmeC2 from Rumen Butyrivibrio .
Int J Mol Sci, 24, 2023
|
|
7CWQ
 
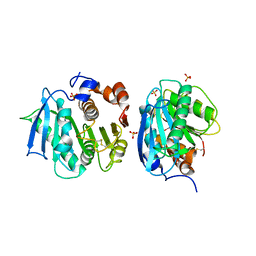 | | Crystal structure of a novel cutinase from Burkhoderiales bacterium RIFCSPLOWO2_02_FULL_57_36 | | Descriptor: | DLH domain-containing protein, SULFATE ION | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2020-08-30 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | General features to enhance enzymatic activity of poly(ethylene terephthalate) hydrolysis.
Nat Catal, 4, 2021
|
|
1QZN
 
 | | Crystal Structure Analysis of a type II cohesin domain from the cellulosome of Acetivibrio cellulolyticus | | Descriptor: | cellulosomal scaffoldin adaptor protein B | | Authors: | Frolow, F, Noach, I, Rosenheck, S, Lamed, R, Qi, X, Shimon, L.J.W, Bayer, E.A. | | Deposit date: | 2003-09-17 | | Release date: | 2004-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a type-II cohesin module from the Bacteroides cellulosolvens cellulosome reveals novel and distinctive secondary structural elements.
J.Mol.Biol., 348, 2005
|
|
5KWS
 
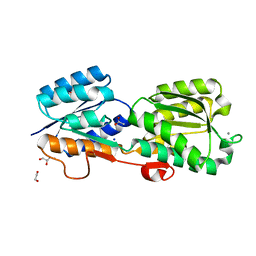 | | Crystal Structure of Galactose Binding Protein from Yersinia pestis in the Complex with beta D Glucose | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-19 | | Release date: | 2016-08-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.316 Å) | | Cite: | Crystal Structure of Galactose Binding Protein from Yersinia pestis in the Complex with beta D Glucose
To Be Published
|
|
4V50
 
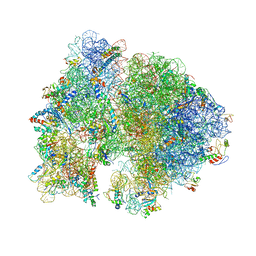 | | Crystal Structure of Ribosome with messenger RNA and the Anticodon stem-loop of P-site tRNA. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Berk, V, Zhang, W, Pai, R.D, Cate, J.H.D. | | Deposit date: | 2006-08-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural basis for mRNA and tRNA positioning on the ribosome.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1QNH
 
 | |
1QK3
 
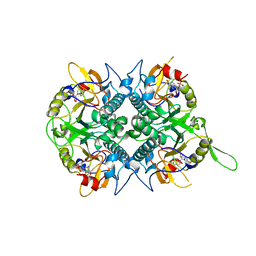 | | TOXOPLASMA GONDII HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE GMP COMPLEX | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE | | Authors: | Heroux, A, White, E.L, Ross, L.J, Borhani, D.W. | | Deposit date: | 1999-07-09 | | Release date: | 1999-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of the Toxoplasma Gondii Hypoxanthine-Guanine Phosphoribosyltransferase Gmp and -Imp Complexes: Comparison of Purine Binding Interactions with the Xmp Complex
Biochemistry, 38, 1999
|
|
1QK5
 
 | | TOXOPLASMA GONDII HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE WITH XMP, PYROPHOSPHATE AND TWO MG2+ IONS | | Descriptor: | HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Heroux, A, White, E.L, Ross, L.J, Davis, R.L, Borhani, D.W. | | Deposit date: | 1999-07-09 | | Release date: | 1999-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Toxoplasma Gondii Hypoxanthine-Guanine Phosphoribosyltransferase with Xmp, Pyrophosphate and Two Mg2+ Ions Bound: Insights Into the Catalytic Mechanism
Biochemistry, 38, 1999
|
|
