7OKF
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 8c | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[4-(4-methylpiperazin-1-yl)-2-oxidanylidene-1H-quinolin-6-yl]amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
7OKJ
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 12c and its enantiomer 12b | | Descriptor: | (2R)-2-[[6-[(2-chloranyl-3-cyano-pyridin-4-yl)amino]-2-oxidanylidene-1H-quinolin-4-yl]amino]-N-methyl-propanamide, (2S)-2-[[6-[(2-chloranyl-3-cyano-pyridin-4-yl)amino]-2-oxidanylidene-1H-quinolin-4-yl]amino]-N-methyl-propanamide, 1,2-ETHANEDIOL, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
6JKI
 
 | | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Jaroensuk, J, Liew, C.W, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Zhong, W.H, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2019-03-01 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD fromPseudomonas aeruginosa.
Rna, 25, 2019
|
|
5Z53
 
 | | Crystal structure of a cyclase Filc from Fischerella sp. in complex with cyclo-L-Arg-D-Pro | | Descriptor: | 12-epi-hapalindole U synthase, CALCIUM ION, SULFATE ION, ... | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-01-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7OKE
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[(1-methyl-2-oxidanylidene-quinolin-6-yl)amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
7OKI
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 12b | | Descriptor: | (2R)-2-[[6-[(2-chloranyl-3-cyano-pyridin-4-yl)amino]-2-oxidanylidene-1H-quinolin-4-yl]amino]-N-methyl-propanamide, 1,2-ETHANEDIOL, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
8WCP
 
 | |
7OKH
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 8f | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[4-(ethylamino)-2-oxidanylidene-1H-quinolin-6-yl]amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
1TEC
 
 | | CRYSTALLOGRAPHIC REFINEMENT BY INCORPORATION OF MOLECULAR DYNAMICS. THE THERMOSTABLE SERINE PROTEASE THERMITASE COMPLEXED WITH EGLIN-C | | Descriptor: | CALCIUM ION, EGLIN C, SODIUM ION, ... | | Authors: | Gros, P, Dijkstra, B.W, Hol, W.G.J. | | Deposit date: | 1989-05-24 | | Release date: | 1989-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic refinement by incorporation of molecular dynamics: thermostable serine protease thermitase complexed with eglin c.
Acta Crystallogr.,Sect.B, 45, 1989
|
|
5K2A
 
 | | 2.5 angstrom A2a adenosine receptor structure with sulfur SAD phasing using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 chimera, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
7RBT
 
 | | cryo-EM structure of human Gastric inhibitory polypeptide receptor GIPR bound to tirzepatide | | Descriptor: | 2-fluoro-4-[(1R)-6-methoxy-1-methyl-2-{(1S)-1-[4-(propan-2-yl)phenyl]ethyl}-1,2,3,4-tetrahydroisoquinolin-5-yl]-6-[(2-methylpropyl)amino]phenol, Gastric inhibitory polypeptide receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Sun, B, Kobilka, B.K, Sloop, K.W, Feng, D, Kobilka, T.S. | | Deposit date: | 2021-07-06 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural determinants of dual incretin receptor agonism by tirzepatide.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3P0U
 
 | | Crystal Structure of the ligand binding domain of human testicular receptor 4 | | Descriptor: | Nuclear receptor subfamily 2 group C member 2 | | Authors: | Zhou, X.E, Suino-Powell, K.M, Xu, Y, Chan, C.-W, Kruse, S.W, Reynolds, R, Engel, J.D, Xu, H.E. | | Deposit date: | 2010-09-29 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Orphan Nuclear Receptor TR4 Is a Vitamin A-activated Nuclear Receptor.
J.Biol.Chem., 286, 2011
|
|
7RG9
 
 | | cryo-EM of human Glucagon-like peptide 1 receptor GLP-1R in apo form | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, B, Kobilka, B.K, Sloop, K.W, Feng, D, Kobilka, T.S. | | Deposit date: | 2021-07-14 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural determinants of dual incretin receptor agonism by tirzepatide.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RA3
 
 | | cryo-EM of human Gastric inhibitory polypeptide receptor GIPR bound to GIP | | Descriptor: | Gastric inhibitory polypeptide, Gastric inhibitory polypeptide receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Sun, B, Kobilka, B.K, Sloop, K.W, Feng, D, Kobilka, T.S. | | Deposit date: | 2021-06-29 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural determinants of dual incretin receptor agonism by tirzepatide.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2W4M
 
 | | The Crystal Structure of human N-acetylneuraminic acid phosphatase, NANP | | Descriptor: | CHLORIDE ION, N-ACYLNEURAMINATE-9-PHOSPHATASE, PHOSPHATE ION, ... | | Authors: | Ugochukwu, E, Cocking, R, Yue, W.W, Pike, A.C.W, Roos, A, Muniz, J.R.C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Oppermann, U. | | Deposit date: | 2008-11-28 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of Human N-Acetylneuraminic Acid Phosphatase, Nanp
To be Published
|
|
4ELH
 
 | | Structure-activity relationship guides enantiomeric preference among potent inhibitors of B. anthracis dihydrofolate reductase | | Descriptor: | (2E)-3-{5-[(2,4-diaminopyrimidin-5-yl)methyl]-2,3-dimethoxyphenyl}-1-[(1R)-1-(2-methylprop-1-en-1-yl)phthalazin-2(1H)-y l]prop-2-en-1-one, (2E)-3-{5-[(2,4-diaminopyrimidin-5-yl)methyl]-2,3-dimethoxyphenyl}-1-[(1S)-1-(2-methylprop-1-en-1-yl)phthalazin-2(1H)-y l]prop-2-en-1-one, CALCIUM ION, ... | | Authors: | Bourne, C.R, Barrow, W.W. | | Deposit date: | 2012-04-10 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Structure-activity relationship for enantiomers of potent inhibitors of B. anthracis dihydrofolate reductase.
Biochim.Biophys.Acta, 1834, 2013
|
|
1TLA
 
 | | HYDROPHOBIC CORE REPACKING AND AROMATIC-AROMATIC INTERACTION IN THE THERMOSTABLE MUTANT OF T4 LYSOZYME SER 117 (RIGHT ARROW) PHE | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, T4 LYSOZYME | | Authors: | Anderson, D.E, Hurley, J.H, Nicholson, H, Baase, W.A, Matthews, B.W. | | Deposit date: | 1993-03-22 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hydrophobic core repacking and aromatic-aromatic interaction in the thermostable mutant of T4 lysozyme Ser 117-->Phe.
Protein Sci., 2, 1993
|
|
6E0E
 
 | | Crystal structure of Glucokinase in complex with compound 6 | | Descriptor: | 2-({2-[(4-methyl-1,3-thiazol-2-yl)amino]pyridin-3-yl}oxy)benzonitrile, Glucokinase, alpha-D-glucopyranose | | Authors: | Hinklin, R.J, Baer, B.R, Boyd, S.A, Chicarelli, M.D, Condroski, K.R, DeWolf, W.E, Fischer, J, Frank, M, Hingorani, G.P, Lee, P.A, Neitzel, N.A, Pratt, S.A, Singh, A, Sullivan, F.X, Turner, T, Voegtli, W.C, Wallace, E.M, Williams, L, Aicher, T.D. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and preclinical development of AR453588 as an anti-diabetic glucokinase activator.
Bioorg.Med.Chem., 28, 2020
|
|
3B4O
 
 | | Crystal structure of phenazine biosynthesis protein PhzA/B from Burkholderia cepacia R18194, apo form | | Descriptor: | ACETATE ION, Phenazine biosynthesis protein A/B | | Authors: | Ahuja, E.G, Janning, P, Mentel, M, Graebsch, A, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2007-10-24 | | Release date: | 2008-12-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PhzA/B catalyzes the formation of the tricycle in phenazine biosynthesis.
J.Am.Chem.Soc., 130, 2008
|
|
7RSY
 
 | | HIV-1 gp120 complex with CJF-III-049-R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 gp120 Clade C1086, N~1~-{(1R,2R,3S)-2-(carbamimidamidomethyl)-3-[(3R)-3,4-dihydroxybutyl]-5-[(methylamino)methyl]-2,3-dihydro-1H-inden-1-yl}-N~2~-(4-chloro-3-fluorophenyl)ethanediamide | | Authors: | Liang, S, Hendrickson, W.A. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of gp120 Residue His105 as a Novel Target for HIV-1 Neutralization by Small-Molecule CD4-Mimics.
Acs Med.Chem.Lett., 12, 2021
|
|
6OBS
 
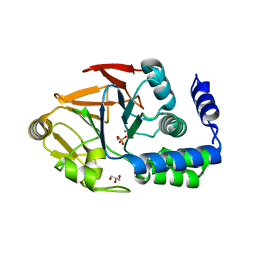 | | PP1 Y134K | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7RSZ
 
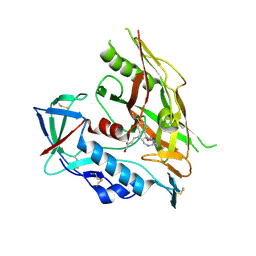 | | HIV-1 gp120 complex with CJF-II-204 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 gp120 Clade C1086, N~1~-{(1R,2R,3S)-2-(carbamimidamidomethyl)-3-[(3R)-3,4-dihydroxybutyl]-5-[(methylamino)methyl]-2,3-dihydro-1H-inden-1-yl}-N~2~-(4-chloro-3-fluorophenyl)ethanediamide, ... | | Authors: | Liang, S, Hendrickson, W.A. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Identification of gp120 Residue His105 as a Novel Target for HIV-1 Neutralization by Small-Molecule CD4-Mimics.
Acs Med.Chem.Lett., 12, 2021
|
|
7RSX
 
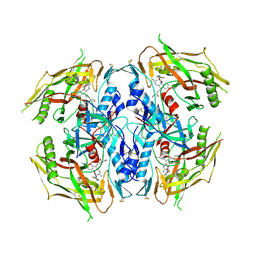 | | HIV-1 gp120 complex with CJF-III-049-S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENVELOPE GLYCOPROTEIN GP120, N~1~-{(1R,2R,3S)-2-(carbamimidamidomethyl)-3-[(3S)-3,4-dihydroxybutyl]-5-[(methylamino)methyl]-2,3-dihydro-1H-inden-1-yl}-N~2~-(4-chloro-3-fluorophenyl)ethanediamide | | Authors: | Liang, S, Hendrickson, W.A. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification of gp120 Residue His105 as a Novel Target for HIV-1 Neutralization by Small-Molecule CD4-Mimics.
Acs Med.Chem.Lett., 12, 2021
|
|
5IJ7
 
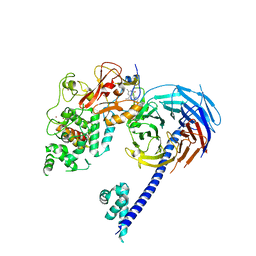 | | Structure of Hs/AcPRC2 in complex with a pyridone inhibitor | | Descriptor: | 5,8-dichloro-2-[(4-ethyl-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-7-({1-[(2R)-2-hydroxypropanoyl]piperidin-4-yl}oxy)-3,4-dihydroisoquinolin-1(2H)-one, Enhancer of Zeste Homolog 2 (EZH2),Histone-lysine N-methyltransferase EZH2, Polycomb protein EED, ... | | Authors: | Gajiwala, K.S, Brooun, A, Deng, Y.-L, Liu, W. | | Deposit date: | 2016-03-01 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance.
Nat Commun, 7, 2016
|
|
7OKG
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 8e | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[4-(1-methylpyrazol-4-yl)-2-oxidanylidene-1H-quinolin-6-yl]amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
