7H4A
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z285782452 (A71EV2A-x0691) | | Descriptor: | DIMETHYL SULFOXIDE, N-methyl-2-(methylsulfonyl)aniline, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease
To Be Published
|
|
7H4O
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with NCL-00024665 (A71EV2A-x1019) | | Descriptor: | 2-(4-bromanyl-2-methoxy-phenyl)ethanoic acid, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease
To Be Published
|
|
7H4J
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z53825020 (A71EV2A-x0875) | | Descriptor: | 1-(ethanesulfonyl)piperidine-4-carboxylic acid, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease
To Be Published
|
|
7H4R
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z1741976468 (A71EV2A-x1081) | | Descriptor: | DIMETHYL SULFOXIDE, Protease 2A, SULFATE ION, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease
To Be Published
|
|
7H4W
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z56042334 (A71EV2A-x1140) | | Descriptor: | 1-(2-chlorophenyl)-2-(pyrrolidin-1-yl)ethan-1-one, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease
To Be Published
|
|
7H53
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with NCL-00024387 (A71EV2A-x1255) | | Descriptor: | 4-bromanyl-1-(2-methoxyethyl)pyridin-2-one, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease
To Be Published
|
|
7H4Y
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z362043378 (A71EV2A-x1146) | | Descriptor: | 2-phenylpyrimidin-5-ol, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease
To Be Published
|
|
7H54
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z57473948 (A71EV2A-x1292) | | Descriptor: | DIMETHYL SULFOXIDE, N-cyclopropyl-2-hydroxybenzamide, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease
To Be Published
|
|
7H52
 
 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z1203730981 (A71EV2A-x1209) | | Descriptor: | 2-bromo-1H-imidazole, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease
To Be Published
|
|
4ZJP
 
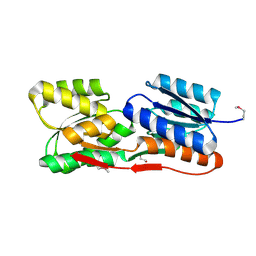 | | Structure of an ABC-Transporter Solute Binding Protein (SBP_IPR025997) from Actinobacillus Succinogenes (Asuc_0197, TARGET EFI-511067) with bound beta-D-ribopyranose | | Descriptor: | 1,2-ETHANEDIOL, Monosaccharide-transporting ATPase, beta-D-ribopyranose | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Glenn, A.S, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure of an ABC-Transporter Solute Binding Protein (SBP_IPR025997) from Actinobacillus Succinogenes (Asuc_0197, TARGET EFI-511067) with bound beta-D-ribopyranose
To be published
|
|
7H3N
 
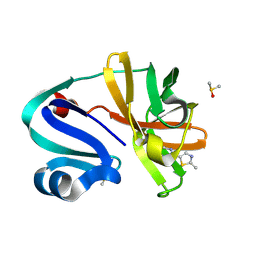 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z851110644 (A71EV2A-x0446) | | Descriptor: | DIMETHYL SULFOXIDE, N,2-dimethyl-1,3-thiazole-5-carboxamide, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease
To Be Published
|
|
7H50
 
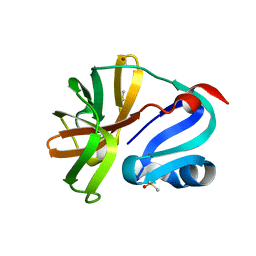 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z1143441220 (A71EV2A-x1169) | | Descriptor: | 1,3-oxazole-4-carbonitrile, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease
To Be Published
|
|
1DAG
 
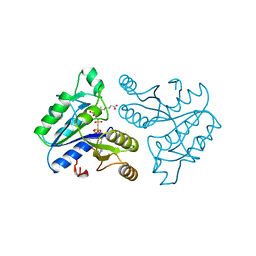 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID AND 5'-ADENOSYL-METHYLENE-TRIPHOSPHATE | | Descriptor: | 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID, DETHIOBIOTIN SYNTHETASE, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
8EZW
 
 | |
4ZN9
 
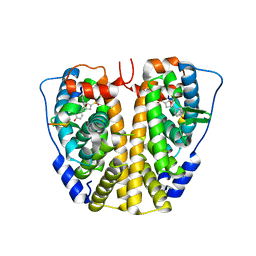 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with Oxabicyclic Heptene Sulfonate (OBHS) | | Descriptor: | Estrogen receptor, Nuclear receptor-interacting peptide, cyclohexa-2,5-dien-1-yl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-04 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.215 Å) | | Cite: | Development of selective estrogen receptor modulator (SERM)-like activity through an indirect mechanism of estrogen receptor antagonism: defining the binding mode of 7-oxabicyclo[2.2.1]hept-5-ene scaffold core ligands.
Chemmedchem, 7, 2012
|
|
2YYJ
 
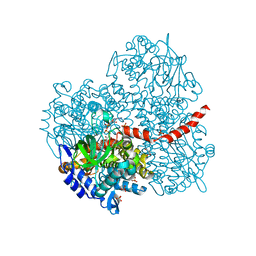 | | Crystal structure of the oxygenase component (HpaB) of 4-hydroxyphenylacetate 3-monooxygenase complexed with FAD and 4-hydroxyphenylacetate | | Descriptor: | 4-HYDROXYPHENYLACETATE, 4-hydroxyphenylacetate-3-hydroxylase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-04-30 | | Release date: | 2007-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
7UQC
 
 | |
8EQ4
 
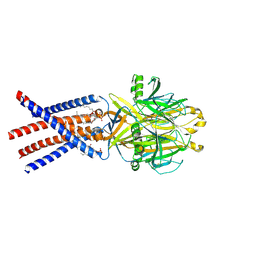 | | Human PAC in nanodisc at pH 4.0 with PI(4,5)P2 diC8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Proton-activated chloride channel, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Ruan, Z, Lu, W. | | Deposit date: | 2022-10-07 | | Release date: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Inhibition of the proton-activated chloride channel PAC by PIP 2.
Elife, 12, 2023
|
|
3D91
 
 | | Human renin in complex with remikiren | | Descriptor: | DIMETHYL SULFOXIDE, Nalpha-[(2S)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-[(1S,2R,3S)-1-(cyclohexylmethyl)-3-cyclopropyl-2,3-dihydroxypropyl]-L-histidinamide, Renin | | Authors: | Prade, L, Bezencon, O, Bur, D, Weller, T, Fischli, W, Remen, L. | | Deposit date: | 2008-05-26 | | Release date: | 2008-06-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human renin in complex with remikiren
to be published
|
|
8ESX
 
 | | HIV protease in complex with benzoxaborolone analog of darunavir | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Windsor, I.W, Graham, B.J, Raines, R.T. | | Deposit date: | 2022-10-15 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Inhibition of HIV-1 Protease by a Boronic Acid with High Oxidative Stability.
Acs Med.Chem.Lett., 14, 2023
|
|
8EMC
 
 | |
7GZ9
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000462-001 | | Descriptor: | Non-structural protein 3, N~2~-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-D-valinamide | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GYZ
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000035-001 | | Descriptor: | N-(4-methyl-3,4-dihydro-2H-1,4-benzoxazin-5-yl)-N'-(1H-pyrazolo[3,4-b]pyridin-5-yl)urea, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.123 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZL
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0008304-001 | | Descriptor: | (3R)-3-[4-(cyclopropylcarbamamido)benzamido]-3-[3-(difluoromethyl)phenyl]propanoic acid, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZ2
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000072-001 | | Descriptor: | (2R)-1-(7H-pyrrolo[2,3-d]pyrimidine-4-carbonyl)azepane-2-carboxylic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.137 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
