2BJ4
 
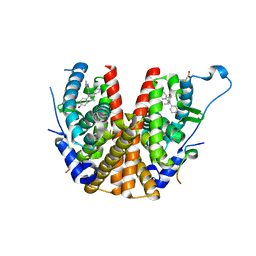 | | ESTROGEN RECEPTOR ALPHA LBD IN COMPLEX WITH A PHAGE-DISPLAY DERIVED PEPTIDE ANTAGONIST | | Descriptor: | 4-HYDROXYTAMOXIFEN, ESTROGEN RECEPTOR, PEPTIDE ANTAGONIST | | Authors: | Kong, E, Heldring, N, Gustafsson, J.A, Treuter, E, Hubbard, R.E, Pike, A.C.W. | | Deposit date: | 2005-01-28 | | Release date: | 2005-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Delineation of a Unique Protein-Protein Interaction Site on the Surface of the Estrogen Receptor
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BSM
 
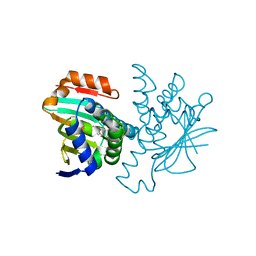 | | Novel, potent small molecule inhibitors of the molecular chaperone Hsp90 discovered through structure-based design | | Descriptor: | 5-(5-CHLORO-2,4-DIHYDROXYPHENYL)-N-ETHYL-4-(4-METHOXYPHENYL)-1H-PYRAZOLE-3-CARBOXAMIDE, HEAT SHOCK PROTEIN HSP90-ALPHA | | Authors: | Dymock, B.W, Barril, X, Brough, P.A, Cansfield, J.E, Massey, A, McDonald, E, Hubbard, R.E, Surgenor, A, Roughley, S.D, Webb, P, Workman, P, Wright, L, Drysdale, M.J. | | Deposit date: | 2005-05-23 | | Release date: | 2005-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Novel, potent small-molecule inhibitors of the molecular chaperone Hsp90 discovered through structure-based design.
J. Med. Chem., 48, 2005
|
|
2BS1
 
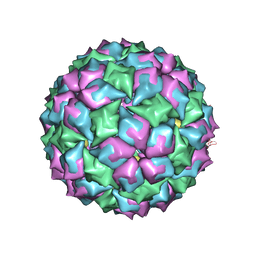 | | MS2 (N87AE89K mutant) - Qbeta RNA hairpin complex | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP *UP*AP*CP*CP*CP*AP*UP*GP*U)-3', MS2 COAT PROTEIN | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-05-13 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of RNA Binding Discrimination between Bacteriophages Qbeta and MS2.
Structure, 14, 2006
|
|
2BT0
 
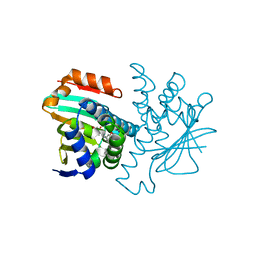 | | Novel, potent small molecule inhibitors of the molecular chaperone Hsp90 discovered through structure-based design | | Descriptor: | 4-[4-(2,3-DIHYDRO-1,4-BENZODIOXIN-6-YL)-3-METHYL-1H-PYRAZOL-5-YL]-6-ETHYLBENZENE-1,3-DIOL, HEAT SHOCK PROTEIN HSP90-ALPHA | | Authors: | Dymock, B.W, Barril, X, Brough, P.A, Cansfield, J.E, Massey, A, McDonald, E, Hubbard, R.E, Surgenor, A, Roughley, S.D, Webb, P, Workman, P, Wright, L, Drysdale, M.J. | | Deposit date: | 2005-05-24 | | Release date: | 2005-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel, potent small-molecule inhibitors of the molecular chaperone Hsp90 discovered through structure-based design.
J. Med. Chem., 48, 2005
|
|
4KSB
 
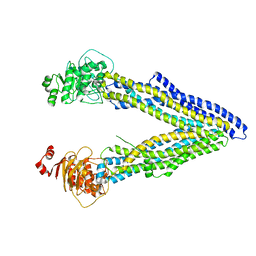 | | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain | | Descriptor: | Multidrug resistance protein 1A | | Authors: | Ward, A, Szewczyk, P, Grimard, V, Lee, C.-W, Martinez, L, Doshi, R, Caya, A, Villaluz, M, Pardon, E, Cregger, C, Swartz, D.J, Falson, P, Urbatsch, I, Govaerts, C, Steyaert, J, Chang, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8001 Å) | | Cite: | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2BT3
 
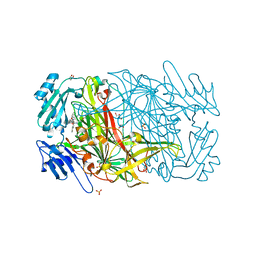 | | AGAO in complex with Ruthenium-C4-wire at 1.73 angstroms | | Descriptor: | BIS[1H,1'H-2,2'-BIPYRIDINATO(2-)-KAPPA~2~N~1~,N~1'~]{3-[4-(1,10-DIHYDRO-1,10-PHENANTHROLIN-4-YL-KAPPA~2~N~1~,N~10~)BUTOXY]-N,N-DIMETHYLANILINATO(2-)}RUTHENIUM, COPPER (II) ION, GLYCEROL, ... | | Authors: | Langley, D.B, Duff, A.P, Freeman, H.C, Guss, J.M, Juda, G.A, Dooley, D.M, Contakes, S.M, Halpern-Manners, N.W, Dunn, A.R, Gray, H.B. | | Deposit date: | 2005-05-26 | | Release date: | 2005-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Reversible Inhibition of Copper Amine Oxidase Activity by Channel-Blocking Ruthenium(II) and Rhenium(I) Molecular Wires.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BZS
 
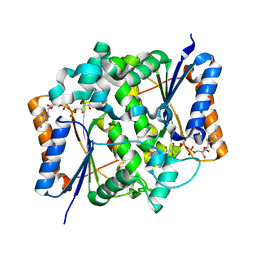 | | Binding of anti-cancer prodrug CB1954 to the activating enzyme NQO2 revealed by the crystal structure of their complex. | | Descriptor: | 5-(AZIRIDIN-1-YL)-2,4-DINITROBENZAMIDE, FLAVIN-ADENINE DINUCLEOTIDE, NRH DEHYDROGENASE [QUINONE] 2, ... | | Authors: | Abu Khader, M.M, Heap, J.T, De Matteis, C, Kellam, B, Doughty, S.W, Minton, N, Paoli, M. | | Deposit date: | 2005-08-22 | | Release date: | 2005-09-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of the Anticancer Prodrug Cb1954 to the Activating Enzyme Nqo2 Revealed by the Crystal Structure of Their Complex.
J.Med.Chem., 48, 2005
|
|
1SGD
 
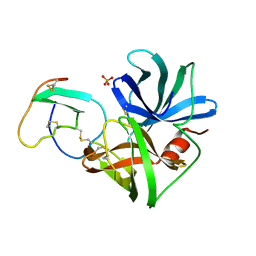 | | ASP 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pH's
To be Published
|
|
5RVW
 
 | | PanDDA analysis group deposition -- Crystal Structure of DHTKD1 in complex with Z1587220559 | | Descriptor: | 2-{[(1H-benzimidazol-2-yl)amino]methyl}phenol, MAGNESIUM ION, Probable 2-oxoglutarate dehydrogenase E1 component DHKTD1, ... | | Authors: | Bezerra, G.A, Foster, W.R, Bailey, H.J, Shrestha, L, Krojer, T, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.614 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
2BII
 
 | | crystal structure of nitrate-reducing fragment of assimilatory nitrate reductase from Pichia angusta | | Descriptor: | (MOLYBDOPTERIN-S,S)-DIOXO-THIO-MOLYBDENUM(IV), GLYCEROL, NITRATE REDUCTASE [NADPH], ... | | Authors: | Fischer, K, Barbier, G, Hecht, H.-J, Mendel, R.R, Campbell, W.H, Schwarz, G. | | Deposit date: | 2005-01-21 | | Release date: | 2005-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Eukaryotic Nitrate Reduction: Crystal Structures of the Nitrate Reductase Active Site
Plant Cell, 17, 2005
|
|
7YSP
 
 | |
2BEP
 
 | | Crystal structure of ubiquitin conjugating enzyme E2-25K | | Descriptor: | BETA-MERCAPTOETHANOL, UBIQUITIN-CONJUGATING ENZYME E2-25 KDA | | Authors: | Pichler, A, Knipscheer, P, Oberhofer, E, Van Dijk, W.J, Korner, R, Velgaard Olsen, J, Jentsch, S, Melchior, F, Sixma, T.K. | | Deposit date: | 2004-11-29 | | Release date: | 2005-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sumo Imodification of the Ubiquitin Conjugating Enzyme E2-25K
Nat.Struct.Mol.Biol., 12, 2005
|
|
7YSN
 
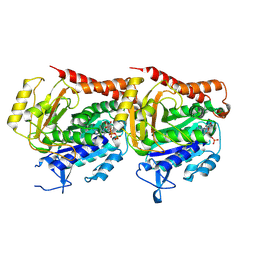 | | Tubulin heterodimer structure of GMPCPP state in solution | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Tubulin alpha-1B chain, ... | | Authors: | Zhou, J, Wang, H.-W. | | Deposit date: | 2022-08-12 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into the mechanism of GTP initiation of microtubule assembly.
Nat Commun, 14, 2023
|
|
7YSQ
 
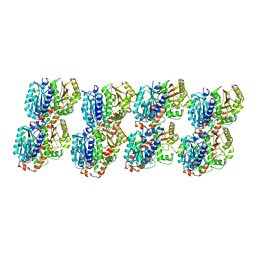 | | GTPgammaS Tube decorated with kinesin | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Tubulin alpha chain, ... | | Authors: | Zhou, J, Wang, H.-W. | | Deposit date: | 2022-08-12 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structural insights into the mechanism of GTP initiation of microtubule assembly.
Nat Commun, 14, 2023
|
|
7YSO
 
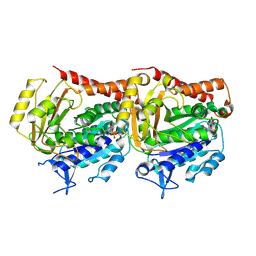 | |
7YSR
 
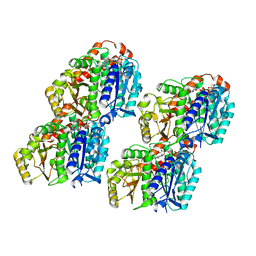 | | GTPgammaS MT decorated with kinesin | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Tubulin alpha chain, ... | | Authors: | Zhou, J, Wang, H.-W. | | Deposit date: | 2022-08-12 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insights into the mechanism of GTP initiation of microtubule assembly.
Nat Commun, 14, 2023
|
|
2BOH
 
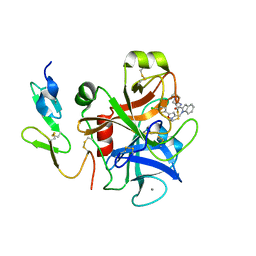 | | Crystal structure of factor Xa in complex with compound "1" | | Descriptor: | 1-{[5-(5-CHLORO-2-THIENYL)ISOXAZOL-3-YL]METHYL}-N-(1-ISOPROPYLPIPERIDIN-4-YL)-1H-INDOLE-2-CARBOXAMIDE, CALCIUM ION, COAGULATION FACTOR XA | | Authors: | Nazare, M, Will, D.W, Matter, H, Schreuder, H, Ritter, K, Urmann, M, Essrich, M, Bauer, A, Wagner, M, Czech, J, Laux, V, Wehner, V. | | Deposit date: | 2005-04-11 | | Release date: | 2006-04-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the Subpockets of Factor Xa Reveals Two Binding Modes for Inhibitors Based on a 2-Carboxyindole Scaffold: A Study Combining Structure-Activity Relationship and X-Ray Crystallography.
J.Med.Chem., 48, 2005
|
|
2BNY
 
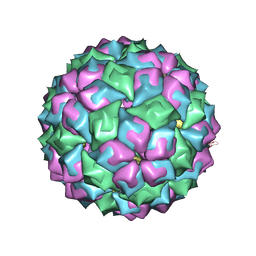 | | MS2 (N87A mutant) - RNA hairpin complex | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP *UP*AP*CP*CP*CP*AP*UP*GP*U)-3', MS2 COAT PROTEIN | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-04-06 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of RNA Binding Discrimination between Bacteriophages Qbeta and MS2.
Structure, 14, 2006
|
|
2BQ7
 
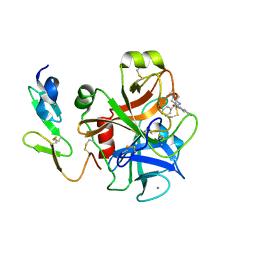 | | Crystal structure of factor Xa in complex with 43 | | Descriptor: | CALCIUM ION, COAGULATION FACTOR X, FACTOR XA, ... | | Authors: | Nazare, M, Will, D.W, Matter, H, Schreuder, H, Ritter, K, Urmann, M, Essrich, M, Bauer, A, Wagner, M, Czech, J, Laux, V, Wehner, V. | | Deposit date: | 2005-04-27 | | Release date: | 2006-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the Subpockets of Factor Xa Reveals Two Binding Modes for Inhibitors Based on a 2-Carboxyindole Scaffold: A Study Combining Structure-Activity Relationship and X-Ray Crystallography.
J.Med.Chem., 48, 2005
|
|
2BIH
 
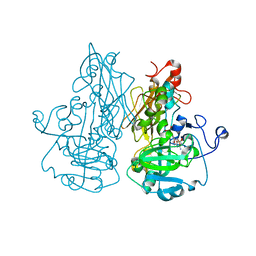 | | crystal structure of the Molybdenum-containing nitrate reducing fragment of Pichia angusta assimilatory nitrate reductase | | Descriptor: | (MOLYBDOPTERIN-S,S)-DIOXO-THIO-MOLYBDENUM(IV), NITRATE REDUCTASE [NADPH] | | Authors: | Fischer, K, Barbier, G, Hecht, H.-J, Mendel, R.R, Campbell, W.H, Schwarz, G. | | Deposit date: | 2005-01-21 | | Release date: | 2005-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of Eukaryotic Nitrate Reduction: Crystal Structures of the Nitrate Reductase Active Site
Plant Cell, 17, 2005
|
|
4KOD
 
 | | Structure of p97 N-D1 R155H mutant in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Xia, D, Tang, W.K. | | Deposit date: | 2013-05-11 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Altered Intersubunit Communication Is the Molecular Basis for Functional Defects of Pathogenic p97 Mutants.
J.Biol.Chem., 288, 2013
|
|
1RYI
 
 | | STRUCTURE OF GLYCINE OXIDASE WITH BOUND INHIBITOR GLYCOLATE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE OXIDASE, GLYCOLIC ACID | | Authors: | Moertl, M, Diederichs, K, Welte, W, Pollegioni, L, Molla, G, Motteran, L, Andriolo, G, Pilone, M.S. | | Deposit date: | 2003-12-22 | | Release date: | 2005-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function correlation in glycine oxidase from Bacillus subtilis
J.Biol.Chem., 279, 2004
|
|
2SGF
 
 | | PHE 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pHs
To be Published
|
|
2BUM
 
 | | Crystal Structure Of Wild-Type Protocatechuate 3,4-Dioxygenase from Acinetobacter Sp. ADP1 | | Descriptor: | FE (III) ION, HYDROXIDE ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, Valley, M.P, D'Argenio, D.A, Ornston, L.N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2005-06-14 | | Release date: | 2006-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biophysical Analyses of Designed and Selected Mutants of Protocatechuate 3,4-Dioxygenase
Annu.Rev.Microbiol., 58, 2004
|
|
2BUW
 
 | | Crystal Structure of Protocatechuate 3,4-Dioxygenase from Acinetobacter Sp. ADP1 Mutant R457S in Complex with 4-Hydroxybenzoate | | Descriptor: | FE (III) ION, P-HYDROXYBENZOIC ACID, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, Valley, M.P, D'Argenio, D.A, Ornston, L.N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2005-06-20 | | Release date: | 2006-10-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Studies of Intradiol Dioxygenases.
Phd Thesis, 2001
|
|
