8G9N
 
 | | Partial DNA elongation subcomplex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA template, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3X1B
 
 | | Crystal structure of laccase from Lentinus sp. at 1.8 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jeng, W.Y, Shyur, L.F, Wang, A.H.J. | | Deposit date: | 2014-10-31 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of laccase from Lentinus sp. at 1.8 A resolution
To be Published
|
|
6SNC
 
 | | crystal structure of LN01 Fab in complex with an HIV-1 gp41 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, LNO1 Heavy Chain, ... | | Authors: | Caillat, C, Pinto, D, Corti, D, Fenwick, C, Pantaleo, G, Weissenhorn, W. | | Deposit date: | 2019-08-23 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for Broad HIV-1 Neutralization by the MPER-Specific Human Broadly Neutralizing Antibody LN01.
Cell Host Microbe, 26, 2019
|
|
8FDT
 
 | | Engineered human dynein motor domain in the microtubule-unbound state with LIS1 complex in the buffer containing ATP-Vi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1,Serine--tRNA ligase, ... | | Authors: | Ton, W, Wang, Y, Chai, P. | | Deposit date: | 2022-12-04 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Microtubule-binding-induced allostery triggers LIS1 dissociation from dynein prior to cargo transport.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1JYW
 
 | |
1RPQ
 
 | | High Affinity IgE Receptor (alpha chain) Complexed with Tight-Binding E131 'zeta' Peptide from Phage Display | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stamos, J, Eigenbrot, C, Nakamura, G.R, Reynolds, M.E, Yin, J.P, Lowman, H.B, Fairbrother, W.J, Starovasnik, M.A. | | Deposit date: | 2003-12-03 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Convergent Recognition of the IgE Binding Site on the High-Affinity IgE Receptor.
Structure, 12, 2004
|
|
8FD6
 
 | | Engineered human dynein motor domain in the microtubule-unbound state in the buffer containing ATP-Vi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1,Serine--tRNA ligase, ... | | Authors: | Ton, W, Wang, Y, Chai, P. | | Deposit date: | 2022-12-02 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Microtubule-binding-induced allostery triggers LIS1 dissociation from dynein prior to cargo transport.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3ZOV
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH CHEMICAL LIGAND | | Descriptor: | 5-(2,2,2-Trifluoro-ethoxy)-pyridine-2-carboxylic acid [3-((S)-2-amino-1,4-dimethyl-6-oxo-1,4,5,6-tetrahydro-pyrimidin-4-yl)-phenyl]-amide, BETA-SECRETASE 1, DIMETHYL SULFOXIDE, ... | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-02-25 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6BOJ
 
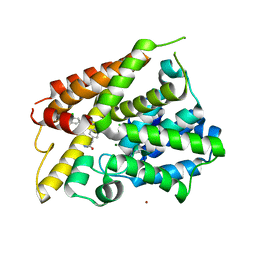 | | Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with BPN5004 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(4-{[2-(3-chlorophenyl)-6-ethylpyrimidin-4-yl]methyl}phenyl)acetamide, CHLORIDE ION, ... | | Authors: | Fox III, D, Fairman, J.W, Gurney, M.E. | | Deposit date: | 2017-11-20 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Memory enhancing effects of BPN14770, an allosteric inhibitor of phosphodiesterase-4D, in wild-type and humanized mice.
Neuropsychopharmacology, 43, 2018
|
|
1NNE
 
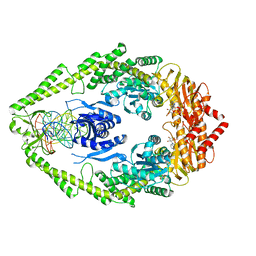 | | Crystal Structure of the MutS-ADPBeF3-DNA complex | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*GP*CP*GP*AP*CP*GP*CP*TP*AP*GP*CP*GP*TP*GP*CP*GP*GP*CP*TP*CP*GP*TP*C)-3', 5'-D(P*GP*GP*AP*CP*GP*AP*GP*CP*CP*GP*CP*CP*GP*CP*TP*AP*GP*CP*GP*TP*CP*G)-3', ... | | Authors: | Alani, E, Lee, J.Y, Schofield, M.J, Kijas, A.W, Hsieh, P, Yang, W. | | Deposit date: | 2003-01-13 | | Release date: | 2003-05-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structure and biochemical analysis of the MutS-ADP-Beryllium Fluoride complex
suggests a conserved mechanism for ATP interactions in mismatch repair
J.Biol.Chem., 278, 2003
|
|
6BQG
 
 | | Crystal structure of 5-HT2C in complex with ergotamine | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562, Ergotamine | | Authors: | Peng, Y, McCorvy, J.D, Harpsoe, K, Lansu, K, Yuan, S, Popov, P, Qu, L, Pu, M, Che, T, Nikolajse, L.F, Huang, X.P, Wu, Y, Shen, L, Bjorn-Yoshimoto, W.E, Ding, K, Wacker, D, Han, G.W, Cheng, J, Katritch, V, Jensen, A.A, Hanson, M.A, Zhao, S, Gloriam, D.E, Roth, B.L, Stevens, R.C, Liu, Z. | | Deposit date: | 2017-11-27 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology.
Cell, 172, 2018
|
|
1RL1
 
 | | Solution structure of human Sgt1 CS domain | | Descriptor: | Suppressor of G2 allele of SKP1 homolog | | Authors: | Lee, Y.-T, Jacob, J, Michowski, W, Nowotny, M, Kuznicki, J, Chazin, W.J. | | Deposit date: | 2003-11-24 | | Release date: | 2004-05-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Human Sgt1 Binds HSP90 through the CHORD-Sgt1 Domain and Not the Tetratricopeptide Repeat Domain
J.Biol.Chem., 279, 2004
|
|
1ROV
 
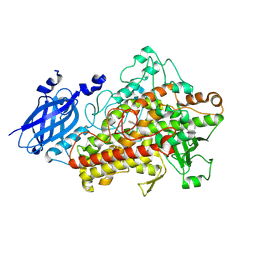 | | Lipoxygenase-3 Treated with Cumene Hydroperoxide | | Descriptor: | FE (III) ION, Seed lipoxygenase-3 | | Authors: | Vahedi-Faridi, A, Brault, P.A, Shah, P, Kim, Y.W, Dunham, W.R, Funk, M.O. | | Deposit date: | 2003-12-02 | | Release date: | 2004-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interaction between non-heme iron of lipoxygenases and cumene hydroperoxide: basis for enzyme activation, inactivation, and inhibition
J.Am.Chem.Soc., 126, 2004
|
|
1HNH
 
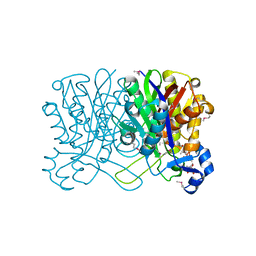 | | CRYSTAL STRUCTURE OF BETA-KETOACYL-ACP SYNTHASE III + DEGRADED FORM OF ACETYL-COA | | Descriptor: | BETA-KETOACYL-ACYL CARRIER PROTEIN SYNTHASE III, COENZYME A | | Authors: | Qiu, X, Janson, C.A, Smith, W.W, Head, M, Lonsdale, J, Konstantinidis, A.K. | | Deposit date: | 2000-12-07 | | Release date: | 2000-12-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Refined structures of beta-ketoacyl-acyl carrier protein synthase III.
J.Mol.Biol., 307, 2001
|
|
1HND
 
 | | CRYSTAL STRUCTURE OF BETA-KETOACYL-ACP SYNTHASE III-COA COMPLEX | | Descriptor: | BETA-KETOACYL-ACYL CARRIER PROTEIN SYNTHASE III, COENZYME A | | Authors: | Qiu, X, Janson, C.A, Smith, W.W, Head, M, Lonsdale, J, Konstantinidis, A.K. | | Deposit date: | 2000-12-07 | | Release date: | 2000-12-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Refined structures of beta-ketoacyl-acyl carrier protein synthase III.
J.Mol.Biol., 307, 2001
|
|
3ZKQ
 
 | | BACE2 XAPERONE COMPLEX | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-SECRETASE 2, CHLORIDE ION, ... | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-01-24 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2FR2
 
 | |
6J6W
 
 | | Mutant K23N of heat shock factor 4-DBD | | Descriptor: | Heat shock factor protein 4, NITRATE ION, SODIUM ION | | Authors: | Xiao, Z.Y, Cui, L.W, Wang, S, Liu, W. | | Deposit date: | 2019-01-16 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | Structure of the mutant K23N of heat shock factor 4-DBD at 1.69 Angstroms resolution.
To Be Published
|
|
1NO6
 
 | | Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor Compound 5 Using a Linked-Fragment Strategy | | Descriptor: | 2-[(CARBOXYCARBONYL)(1-NAPHTHYL)AMINO]BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Szczepankiewicz, B.G, Liu, G, Hajduk, P.J, Abad-Zapatero, C, Pei, Z, Xin, Z, Lubben, T, Trevillyan, J.M, Stashko, M.A, Ballaron, S.J, Liang, H, Huang, F, Hutchins, C.W, Fesik, S.W, Jirousek, M.R. | | Deposit date: | 2003-01-15 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor
Using a Linked-Fragment Strategy
J.Am.Chem.Soc., 125, 2003
|
|
2VV5
 
 | | The open structure of MscS | | Descriptor: | SMALL-CONDUCTANCE MECHANOSENSITIVE CHANNEL | | Authors: | Wang, W, Dong, C, Johnson, K.A, Naismith, J.H. | | Deposit date: | 2008-06-03 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The Structure of an Open Form of an E. Coli Mechanosensitive Channel at 3.45 A Resolution.
Science, 321, 2008
|
|
1HRD
 
 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE | | Authors: | Britton, K.L, Baker, P.J, Stillman, T.J, Rice, D.W. | | Deposit date: | 1996-04-03 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The structure of Pyrococcus furiosus glutamate dehydrogenase reveals a key role for ion-pair networks in maintaining enzyme stability at extreme temperatures.
Structure, 3, 1995
|
|
8G5C
 
 | |
5M7J
 
 | | Blastochloris viridis photosynthetic reaction center structure using best crystal approach | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL A, ... | | Authors: | Sharma, A.S, Johansson, L, Dunevall, E, Wahlgren, W.Y, Neutze, R, Katona, G. | | Deposit date: | 2016-10-28 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Asymmetry in serial femtosecond crystallography data.
Acta Crystallogr A Found Adv, 73, 2017
|
|
1HNJ
 
 | | CRYSTAL STRUCTURE OF BETA-KETOACYL-ACP SYNTHASE III + MALONYL-COA | | Descriptor: | BETA-KETOACYL-ACYL CARRIER PROTEIN SYNTHASE III, MALONYL-COENZYME A, PHOSPHATE ION | | Authors: | Qiu, X, Janson, C.A, Smith, W.W, Head, M, Lonsdale, J, Konstantinidis, A.K. | | Deposit date: | 2000-12-07 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Refined structures of beta-ketoacyl-acyl carrier protein synthase III.
J.Mol.Biol., 307, 2001
|
|
1NW4
 
 | | Crystal Structure of Plasmodium falciparum Purine Nucleoside Phosphorylase in complex with ImmH and Sulfate | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Shi, W, Ting, L.M, Kicska, G.A, Lewandowicz, A, Tyler, P.C, Evans, G.B, Furneaux, R.H, Kim, K, Almo, S.C, Schramm, V.L. | | Deposit date: | 2003-02-05 | | Release date: | 2004-03-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Plasmodium falciparum Purine Nucleoside Phosphorylase: CRYSTAL STRUCTURES, IMMUCILLIN INHIBITORS, AND DUAL CATALYTIC FUNCTION.
J.Biol.Chem., 279, 2004
|
|
