4RFE
 
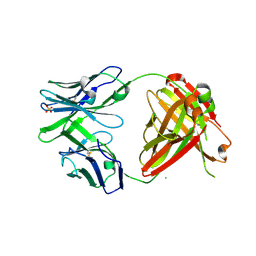 | | Crystal structure of ADCC-potent ANTI-HIV-1 Rhesus macaque antibody JR4 Fab | | 分子名称: | CHLORIDE ION, Fab heavy chain of ADCC-potent anti-HIV-1 antibody JR4, Fab light chain of ADCC-potent anti-HIV-1 antibody JR4, ... | | 著者 | Wu, X, Gohain, N, Tolbert, W.D, Pazgier, M. | | 登録日 | 2014-09-25 | | 公開日 | 2015-07-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Cocrystal Structures of Antibody N60-i3 and Antibody JR4 in Complex with gp120 Define More Cluster A Epitopes Involved in Effective Antibody-Dependent Effector Function against HIV-1.
J.Virol., 89, 2015
|
|
6FUU
 
 | |
8DM5
 
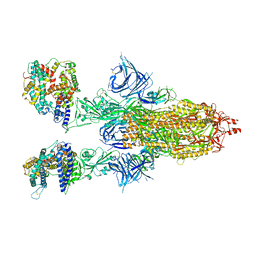 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with human ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | 登録日 | 2022-07-08 | | 公開日 | 2023-02-08 | | 実験手法 | ELECTRON MICROSCOPY (2.51 Å) | | 主引用文献 | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
1U1C
 
 | | Structure of E. coli uridine phosphorylase complexed to 5-benzylacyclouridine (BAU) | | 分子名称: | 1-((2-HYDROXYETHOXY)METHYL)-5-BENZYLPYRIMIDINE-2,4(1H,3H)-DIONE, PHOSPHATE ION, POTASSIUM ION, ... | | 著者 | Bu, W, Settembre, E.C, Ealick, S.E. | | 登録日 | 2004-07-15 | | 公開日 | 2005-07-05 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis for inhibition of Escherichia coli uridine phosphorylase by 5-substituted acyclouridines.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7DWO
 
 | |
4N37
 
 | | Structure of langerin CRD I313 D288 complexed with Me-Man | | 分子名称: | C-type lectin domain family 4 member K, CALCIUM ION, methyl alpha-D-mannopyranoside | | 著者 | Feinberg, H, Rowntree, T.J.W, Tan, S.L.W, Drickamer, K, Weis, W.I, Taylor, M.E. | | 登録日 | 2013-10-06 | | 公開日 | 2013-11-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Common polymorphisms in human langerin change specificity for glycan ligands.
J.Biol.Chem., 288, 2013
|
|
8DWL
 
 | | Inhibitor-3:PP1 coexpressed complex | | 分子名称: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase PPP1R11, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit, ... | | 著者 | Choy, M.S, Srivastava, G, Page, R, Peti, W. | | 登録日 | 2022-08-01 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Inhibitor-3 inhibits Protein Phosphatase 1 via a metal binding dynamic protein-protein interaction.
Nat Commun, 14, 2023
|
|
4MY9
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor C91 | | 分子名称: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, MALONATE ION, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-09-27 | | 公開日 | 2014-06-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5893 Å) | | 主引用文献 | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor C91
To be Published
|
|
7DWN
 
 | | Crystal structure of Vibrio fischeri DarR in complex with DNA reveals the transcriptional activation mechanism of LTTR family members | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Predicted DNA-binding transcriptional regulator | | 著者 | Wang, W.W, Wu, H, He, J.H, Yu, F. | | 登録日 | 2021-01-17 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Crystal structure details of Vibrio fischeri DarR and mutant DarR-M202I from LTTR family reveals their activation mechanism.
Int.J.Biol.Macromol., 183, 2021
|
|
6FPG
 
 | | Structure of the Ustilago maydis chorismate mutase 1 in complex with a Zea mays kiwellin | | 分子名称: | CITRIC ACID, Chromosome 16, whole genome shotgun sequence, ... | | 著者 | Altegoer, F, Steinchen, W, Bange, G. | | 登録日 | 2018-02-09 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A kiwellin disarms the metabolic activity of a secreted fungal virulence factor.
Nature, 565, 2019
|
|
5WUJ
 
 | | Crystal structure of FliF-FliG complex from H. pylori | | 分子名称: | Flagellar M-ring protein, Flagellar motor switch protein FliG, GLYCEROL | | 著者 | Au, S.W, Xue, C, Lam, K.H, Lee, S.H. | | 登録日 | 2016-12-19 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the FliF-FliG complex from Helicobacter pylori yields insight into the assembly of the motor MS-C ring in the bacterial flagellum
J. Biol. Chem., 293, 2018
|
|
8DWK
 
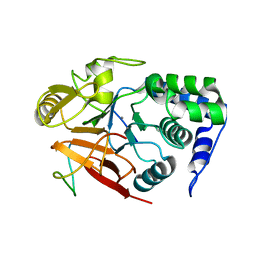 | | Inhibitor-3:PP1 reconstituted complex | | 分子名称: | E3 ubiquitin-protein ligase PPP1R11, MANGANESE (II) ION, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | 著者 | Choy, M.S, Srivastava, G, Page, R, Peti, W. | | 登録日 | 2022-08-01 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Inhibitor-3 inhibits Protein Phosphatase 1 via a metal binding dynamic protein-protein interaction.
Nat Commun, 14, 2023
|
|
6ZHB
 
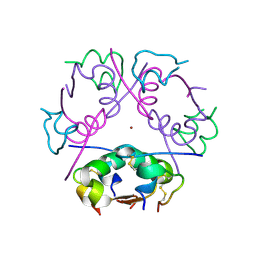 | | 3D electron diffraction structure of bovine insulin | | 分子名称: | Insulin, ZINC ION | | 著者 | Blum, T, Housset, D, Clabbers, M.T.B, van Genderen, E, Bacia-Verloop, M, Zander, U, McCarthy, A.A, Schoehn, G, Ling, W.L, Abrahams, J.P. | | 登録日 | 2020-06-22 | | 公開日 | 2021-01-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (3.25 Å) | | 主引用文献 | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1RKX
 
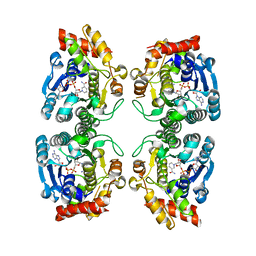 | | Crystal Structure at 1.8 Angstrom of CDP-D-glucose 4,6-dehydratase from Yersinia pseudotuberculosis | | 分子名称: | CDP-glucose-4,6-dehydratase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Vogan, E.M, Bellamacina, C, He, X, Liu, H.W, Ringe, D, Petsko, G.A. | | 登録日 | 2003-11-23 | | 公開日 | 2004-03-30 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure at 1.8 A Resolution of CDP-d-Glucose 4,6-Dehydratase from Yersinia pseudotuberculosis
Biochemistry, 43, 2004
|
|
8JGA
 
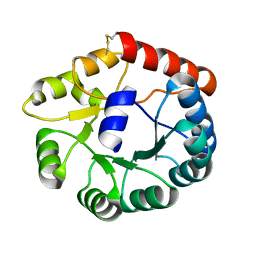 | | Cryo-EM structure of Mi3 fused with FKBP | | 分子名称: | Peptidyl-prolyl cis-trans isomerase FKBP1A,2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase | | 著者 | Zhang, H.W, Kang, W, Xue, C. | | 登録日 | 2023-05-20 | | 公開日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.68 Å) | | 主引用文献 | Dynamic Metabolons Using Stimuli-Responsive Protein Cages.
J.Am.Chem.Soc., 146, 2024
|
|
6ZI8
 
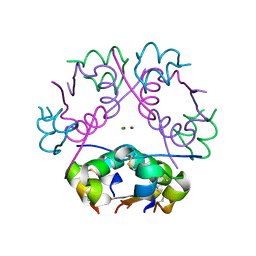 | | X-ray diffraction structure of bovine insulin at 2.3 A resolution | | 分子名称: | CHLORIDE ION, Insulin, ZINC ION | | 著者 | Housset, D, Ling, W.L, Bacia-Verloop, M, Zander, U, McCarthy, A.A, Schoehn, G. | | 登録日 | 2020-06-25 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5W79
 
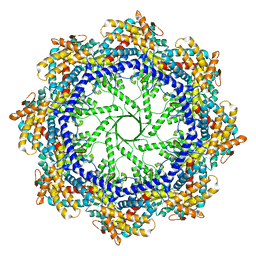 | | Crystal Structure of the Group II Chaperonin from Methanococcus Maripaludis, Cysteine-less mutant in the Apo State | | 分子名称: | Chaperonin, SULFATE ION | | 著者 | Dalton, K.M, Lopez, T, Liu, C, Ralston, C.Y, Pereira, J.H, Chartron, J.W, McAndrew, R.P, Douglas, N.R, Adams, P.D, Pande, V.S, Frydman, J. | | 登録日 | 2017-06-19 | | 公開日 | 2018-06-20 | | 最終更新日 | 2018-08-29 | | 実験手法 | X-RAY DIFFRACTION (3.122 Å) | | 主引用文献 | The Conformational Cycle of the Group II Chaperonin Termini
To Be Published
|
|
8DNL
 
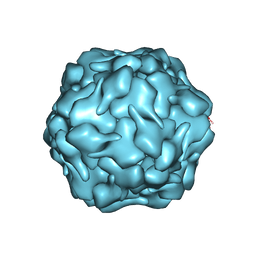 | |
4RC8
 
 | |
8DNA
 
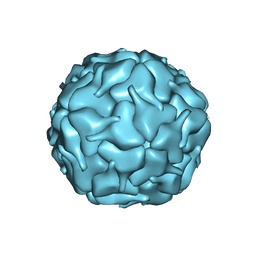 | |
5WGM
 
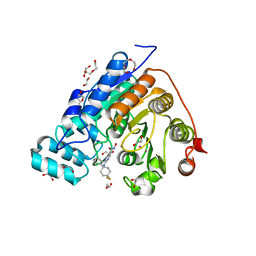 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 in complex with ACY-1083 | | 分子名称: | 1,2-ETHANEDIOL, 2-[(4,4-difluoro-1-phenylcyclohexyl)amino]-N-hydroxypyrimidine-5-carboxamide, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Porter, N.J, Christianson, D.W. | | 登録日 | 2017-07-14 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Unusual zinc-binding mode of HDAC6-selective hydroxamate inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4RZC
 
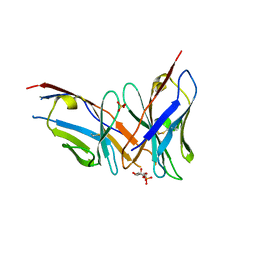 | | Fv M6P-1 in complex with mannose-6-phosphate | | 分子名称: | 6-O-phosphono-alpha-D-mannopyranose, Fv M6P-1 heavy chain, Fv M6P-1 light chain, ... | | 著者 | Blackler, R.J, Evans, D.W, Evans, S.V, Muller-Loennies, S. | | 登録日 | 2014-12-19 | | 公開日 | 2015-11-04 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.723 Å) | | 主引用文献 | Single-chain antibody-fragment M6P-1 possesses a mannose 6-phosphate monosaccharide-specific binding pocket that distinguishes N-glycan phosphorylation in a branch-specific manner.
Glycobiology, 26, 2016
|
|
8DN9
 
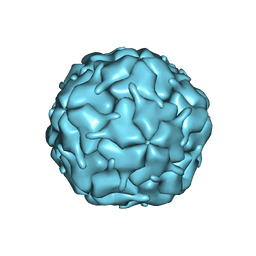 | |
6ZCK
 
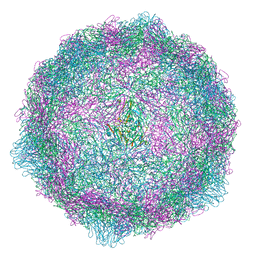 | | Coxsackievirus B4 in complex with capsid binder compound 48 | | 分子名称: | 4-[(6-propoxynaphthalen-2-yl)sulfonylamino]benzoic acid, Capsid protein VP1, Capsid protein VP2, ... | | 著者 | Flatt, J.W, Domanska, A, Butcher, S.J. | | 登録日 | 2020-06-11 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Identification of a conserved virion-stabilizing network inside the interprotomer pocket of enteroviruses.
Commun Biol, 4, 2021
|
|
6ZCL
 
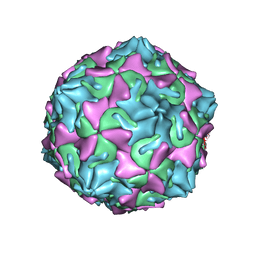 | | Coxsackievirus B3 in complex with capsid binder compound 17 | | 分子名称: | 4-[[4-[1,3-bis(oxidanylidene)isoindol-2-yl]phenyl]sulfonylamino]benzoic acid, MYRISTIC ACID, capsid protein VP1, ... | | 著者 | Domanska, A, Flatt, J.W, Butcher, S.J. | | 登録日 | 2020-06-11 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Identification of a conserved virion-stabilizing network inside the interprotomer pocket of enteroviruses.
Commun Biol, 4, 2021
|
|
