7SDC
 
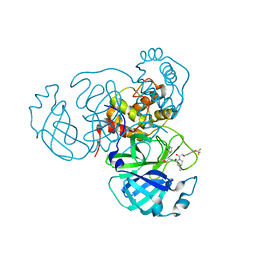 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MI-09 | | 分子名称: | (1R,2S,5S)-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-{[4-(trifluoromethoxy)phenoxy]acetyl}-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Yang, K.S, Liu, W.R. | | 登録日 | 2021-09-29 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | A Systematic Survey of Reversibly Covalent Dipeptidyl Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 66, 2023
|
|
1QKM
 
 | |
1A6B
 
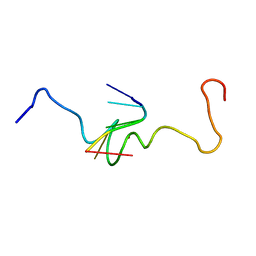 | | NMR STRUCTURE OF THE COMPLEX BETWEEN THE ZINC FINGER PROTEIN NCP10 OF MOLONEY MURINE LEUKEMIA VIRUS AND A SEQUENCE OF THE PSI-PACKAGING DOMAIN OF HIV-1, 20 STRUCTURES | | 分子名称: | DNA (5'-D(*AP*CP*GP*CP*C)-3'), ZINC FINGER PROTEIN NCP10, ZINC ION | | 著者 | Schueler, W, Dong, C.-Z, Wecker, K, Roques, B.P. | | 登録日 | 1998-02-23 | | 公開日 | 1999-08-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the complex between the zinc finger protein NCp10 of Moloney murine leukemia virus and the single-stranded pentanucleotide d(ACGCC): comparison with HIV-NCp7 complexes.
Biochemistry, 38, 1999
|
|
4BDI
 
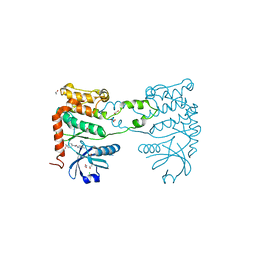 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | 分子名称: | 1,2-ETHANEDIOL, 1-acetyl-N-(5-methylpyridin-2-yl)piperidine-4-carboxamide, CHLORIDE ION, ... | | 著者 | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | 登録日 | 2012-10-05 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
1AAB
 
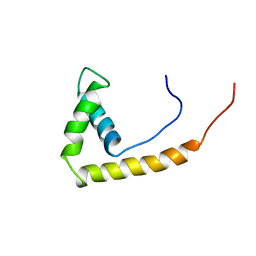 | | NMR STRUCTURE OF RAT HMG1 HMGA FRAGMENT | | 分子名称: | HIGH MOBILITY GROUP PROTEIN | | 著者 | Hardman, C.H, Broadhurst, R.W, Raine, A.R.C, Grasser, K.D, Thomas, J.O, Laue, E.D. | | 登録日 | 1995-10-28 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the A-domain of HMG1 and its interaction with DNA as studied by heteronuclear three- and four-dimensional NMR spectroscopy.
Biochemistry, 34, 1995
|
|
6ISM
 
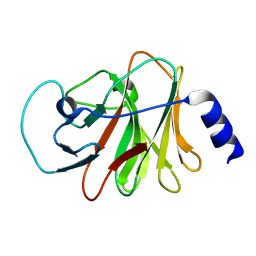 | | Crystal structure of intracellular B30.2 domain of BTN3A1 mutant | | 分子名称: | Butyrophilin subfamily 3 member A1 | | 著者 | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | 登録日 | 2018-11-16 | | 公開日 | 2019-04-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
2R8Y
 
 | | Crystal structure of YrbI phosphatase from Escherichia coli in a complex with Ca | | 分子名称: | CALCIUM ION, CHLORIDE ION, YrbI from Escherichia coli | | 著者 | Tsodikov, O.V, Aggarwal, P, Rubin, J.R, Stuckey, J.A, Woodard, R.W, Biswas, T. | | 登録日 | 2007-09-11 | | 公開日 | 2008-09-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The Tail of KdsC: CONFORMATIONAL CHANGES CONTROL THE ACTIVITY OF A HALOACID DEHALOGENASE SUPERFAMILY PHOSPHATASE.
J.Biol.Chem., 284, 2009
|
|
1A8M
 
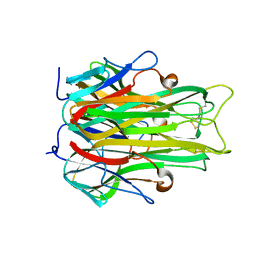 | | TUMOR NECROSIS FACTOR ALPHA, R31D MUTANT | | 分子名称: | TUMOR NECROSIS FACTOR ALPHA | | 著者 | Reed, C, Fu, Z.-Q, Wu, J, Xue, Y.-N, Harrison, R.W, Chen, M.-J, Weber, I.T. | | 登録日 | 1998-03-27 | | 公開日 | 1998-06-17 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of TNF-alpha mutant R31D with greater affinity for receptor R1 compared with R2.
Protein Eng., 10, 1997
|
|
4BDB
 
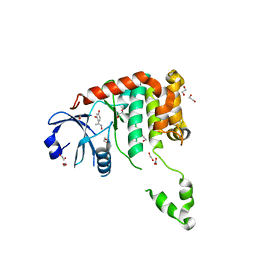 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | 分子名称: | 1,2-ETHANEDIOL, 4-[(E)-C-methyl-N-oxidanyl-carbonimidoyl]benzene-1,3-diol, NITRATE ION, ... | | 著者 | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | 登録日 | 2012-10-05 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
1SAN
 
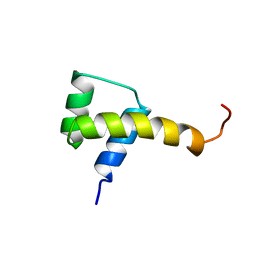 | |
3HHL
 
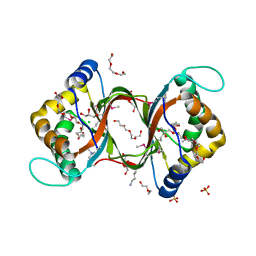 | | Crystal structure of methylated RPA0582 protein | | 分子名称: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Sledz, P, Niedzialkowska, E, Chruszcz, M, Porebski, P, Yim, V, Kudritska, M, Zimmerman, M.D, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-05-15 | | 公開日 | 2009-07-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Crystal structure of methylated RPA0582 protein
To be Published
|
|
4ZUO
 
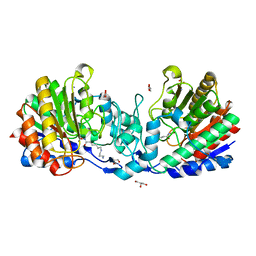 | | Crystal structure of acetylpolyamine amidohydrolase from Mycoplana ramosa in complex with a hydroxamate inhibitor | | 分子名称: | 6-[(3-aminopropyl)amino]-N-hydroxyhexanamide, AMMONIUM ION, Acetylpolyamine aminohydrolase, ... | | 著者 | Decroos, C, Christianson, D.W. | | 登録日 | 2015-05-17 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Design, Synthesis, and Evaluation of Polyamine Deacetylase Inhibitors, and High-Resolution Crystal Structures of Their Complexes with Acetylpolyamine Amidohydrolase.
Biochemistry, 54, 2015
|
|
7YRG
 
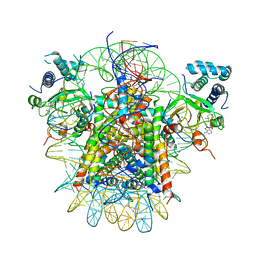 | | histone methyltransferase | | 分子名称: | DNA (146-MER), Histone H2A.Z, Histone H2B 1.1, ... | | 著者 | Li, H, Wang, W.Y. | | 登録日 | 2022-08-09 | | 公開日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1.
Mol.Cell, 83, 2023
|
|
1EAA
 
 | |
6IME
 
 | | Rv2361c complex with substrate analogues | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-methylbut-3-enylsulfanyl(phosphonooxy)phosphinic acid, CARBONATE ION, ... | | 著者 | Ko, T.-P, Guo, R.-T, Chen, C.-C, Liu, W. | | 登録日 | 2018-10-22 | | 公開日 | 2019-09-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Substrate-analogue complex structure of Mycobacterium tuberculosis decaprenyl diphosphate synthase.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
1SBN
 
 | |
7SH9
 
 | |
1EAE
 
 | |
7SH8
 
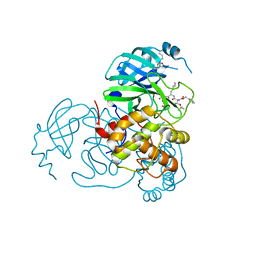 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI88 | | 分子名称: | 3C-like proteinase nsp5, benzyl [(2S,3R)-1-{[(2S)-1-(2-acetyl-2-{[(3S)-2-oxopyrrolidin-3-yl]methyl}hydrazinyl)-3-cyclohexyl-1-oxopropan-2-yl]amino}-3-tert-butoxy-1-oxobutan-2-yl]carbamate (non-preferred name) | | 著者 | Yang, K.S, Liu, W.R. | | 登録日 | 2021-10-08 | | 公開日 | 2023-04-12 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of the SARS-CoV-2 main protease in complex with inhibitors
To Be Published
|
|
1B1B
 
 | | IRON DEPENDENT REGULATOR | | 分子名称: | PROTEIN (IRON DEPENDENT REGULATOR), SULFATE ION, ZINC ION | | 著者 | Pohl, E, Holmes, R.K, Hol, W.G. | | 登録日 | 1998-11-19 | | 公開日 | 1999-12-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of the iron-dependent regulator (IdeR) from Mycobacterium tuberculosis shows both metal binding sites fully occupied.
J.Mol.Biol., 285, 1999
|
|
1OZH
 
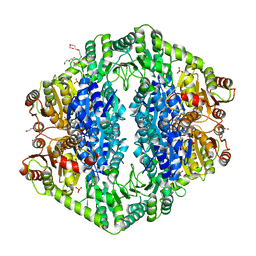 | | The crystal structure of Klebsiella pneumoniae acetolactate synthase with enzyme-bound cofactor and with an unusual intermediate. | | 分子名称: | 2-HYDROXYETHYL DIHYDROTHIACHROME DIPHOSPHATE, Acetolactate synthase, catabolic, ... | | 著者 | Pang, S.S, Duggleby, R.G, Schowen, R.L, Guddat, L.W. | | 登録日 | 2003-04-09 | | 公開日 | 2003-11-04 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Crystal Structures of Klebsiella pneumoniae Acetolactate Synthase with Enzyme-bound Cofactor and with an Unusual Intermediate.
J.Biol.Chem., 279, 2004
|
|
1B4J
 
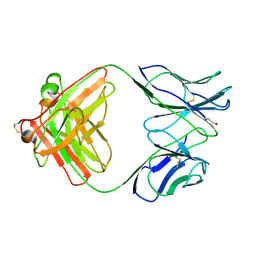 | | COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF A HUMANIZED AND A CHIMERIC FAB OF AN ANTI-GAMMA-INTERFERON ANTIBODY | | 分子名称: | ANTIBODY | | 著者 | Fan, Z, Shan, L, Goldsteen, B.Z, Guddat, L.W, Thakur, A, Landolfi, N.F, Co, M.S, Vasques, M, Queen, C, Ramsland, P.A, Edmundson, A.B. | | 登録日 | 1998-12-22 | | 公開日 | 1999-06-15 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Comparison of the three-dimensional structures of a humanized and a chimeric Fab of an anti-gamma-interferon antibody.
J.Mol.Recog., 12, 1999
|
|
5E04
 
 | |
7SHB
 
 | |
3ERP
 
 | | Structure of IDP01002, a putative oxidoreductase from and essential gene of Salmonella typhimurium | | 分子名称: | 1,2-ETHANEDIOL, CACODYLATE ION, CHLORIDE ION, ... | | 著者 | Singer, A.U, Minasov, G, Evdokimova, E, Brunzelle, J.S, Kudritska, M, Edwards, A.M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2008-10-02 | | 公開日 | 2008-11-04 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural and biochemical studies of novel aldo-keto reductases for the biocatalytic conversion of 3-hydroxybutanal to 1,3-butanediol.
Appl.Environ.Microbiol., 2017
|
|
