9ATS
 
 | | Crystal structure of MERS 3CL protease in complex with a methylcyclohexyl 2-pyrrolidone inhibitor (S-enantiomer) | | 分子名称: | (1S,2S)-2-{[N-({[(2S)-1-(cyclohexylmethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2024-02-27 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
7S4J
 
 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.16 Angstrom resolution | | 分子名称: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | 著者 | Koo, C.W, Rosenzweig, A.C. | | 登録日 | 2021-09-09 | | 公開日 | 2022-03-30 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.16 Å) | | 主引用文献 | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
8YJM
 
 | | Structure of human SPT16 MD-CTD and MCM2 HBD chaperoning a histone H3-H4 tetramer and a single chain H2B-H2A chimera | | 分子名称: | DNA replication licensing factor MCM2, FACT complex subunit SPT16, Histone H2B 1/2/3/4/6,Histone H2A type 1-D, ... | | 著者 | Gan, S.L, Yang, W.S, Xu, R.M. | | 登録日 | 2024-03-02 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (4.15 Å) | | 主引用文献 | Structure of a histone hexamer bound by the chaperone domains of SPT16 and MCM2.
Sci China Life Sci, 67, 2024
|
|
9AT6
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a methylbicyclo[2.2.1]heptene 2-pyrrolidone inhibitor | | 分子名称: | (1R,2S)-2-{[N-({[(2S)-1-{[(1R,2S,4R)-bicyclo[2.2.1]hept-5-en-2-yl]methyl}-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(2S)-1-{[(1R,2S,4R)-bicyclo[2.2.1]hept-5-en-2-yl]methyl}-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | 著者 | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2024-02-26 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
8WTE
 
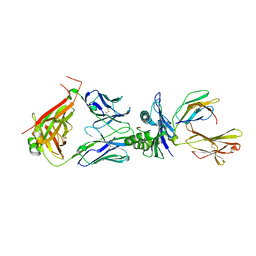 | | Crystal structure of TCR in complex with HLA-A*11:01 bound to KRAS-G12V peptide (VVGAVGVGK) | | 分子名称: | Beta-2-microglobulin, KRAS-G12V nonamer peptide, MHC class I antigen (Fragment), ... | | 著者 | Zhang, M.Y, Luo, L.J, Xu, W, Guan, F.H, Wang, X.Y, Zhu, P, Zhang, J.H, Zhou, X.Y, Wang, F, Ye, S. | | 登録日 | 2023-10-18 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Identification and affinity enhancement of T-cell receptor targeting a KRAS G12V cancer neoantigen.
Commun Biol, 7, 2024
|
|
7S4H
 
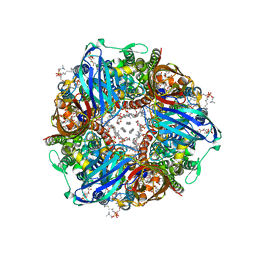 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.14 Angstrom resolution | | 分子名称: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | 著者 | Koo, C.W, Rosenzweig, A.C. | | 登録日 | 2021-09-08 | | 公開日 | 2022-03-30 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.14 Å) | | 主引用文献 | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
8VE1
 
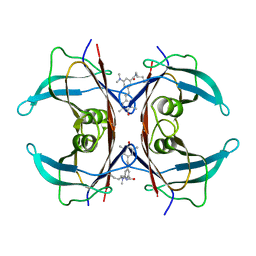 | | Human transthyretin covalently modified with A2-derived stilbene in the canonical conformation | | 分子名称: | 3-(dimethylamino)-5-[(E)-2-(4-hydroxy-3,5-dimethylphenyl)ethenyl]benzoic acid, Transthyretin | | 著者 | Basanta, B, Nugroho, K, Yan, N, Kline, G.M, Tsai, F.J, Wu, M, Kelly, J.W, Lander, G.C. | | 登録日 | 2023-12-18 | | 公開日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | The conformational landscape of human transthyretin revealed by cryo-EM
To Be Published
|
|
9BUL
 
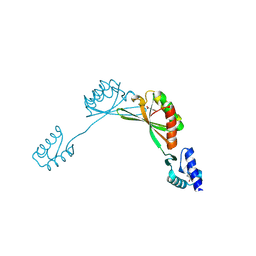 | |
9ASW
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a m-fluorobenzyl 2-pyrrolidone inhibitor | | 分子名称: | (1R,2S)-2-({N-[({(2S)-1-[(3-fluorophenyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(3-fluorophenyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | 著者 | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2024-02-26 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9ASV
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a benzyl 2-pyrrolidone inhibitor | | 分子名称: | (1R,2S)-2-{[N-({[(2S)-1-benzyl-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(2S)-1-benzyl-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | 著者 | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2024-02-26 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
7S4M
 
 | |
6DB7
 
 | |
9CE9
 
 | |
1QS7
 
 | |
6DBO
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | 分子名称: | CALCIUM ION, Forward strand of substrate RSS DNA, Recombination activating gene 1 - MBP chimera, ... | | 著者 | Wu, H, Liao, M, Ru, H, Mi, W. | | 登録日 | 2018-05-03 | | 公開日 | 2018-08-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4W4R
 
 | | Crystal structure of ent-kaurene synthase BJKS from bradyrhizobium japonicum | | 分子名称: | Uncharacterized protein blr2150 | | 著者 | Liu, W, Zheng, Y, Huang, C.H, Ko, T.P, Guo, R.T. | | 登録日 | 2014-08-15 | | 公開日 | 2015-01-14 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structure, function and inhibition of ent-kaurene synthase from Bradyrhizobium japonicum.
Sci Rep, 4, 2014
|
|
9ATD
 
 | | Crystal structure of MERS 3CL protease in complex with a ethylcyclohexyl 2-pyrrolidone inhibitor (S-enantiomer) inhibitor | | 分子名称: | (1R,2S)-2-{[N-({[(2S)-1-(2-cyclohexylethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(2S)-1-(2-cyclohexylethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2024-02-26 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
7S4L
 
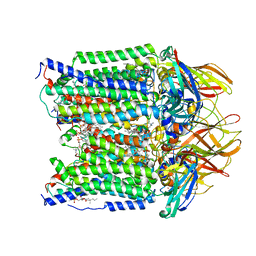 | | CryoEM structure of Methylotuvimicrobium alcaliphilum 20Z pMMO in a POPC nanodisc at 2.46 Angstrom resolution | | 分子名称: | (S)-2,3-bis(hexanoyloxy)propyl(2-(trimethylammonio)ethyl)phosphate, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, COPPER (II) ION, ... | | 著者 | Koo, C.W, Rosenzweig, A.C. | | 登録日 | 2021-09-09 | | 公開日 | 2022-03-30 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.46 Å) | | 主引用文献 | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
9CYS
 
 | | Toxin/immunity complex for a T6SS lipase effector from E. cloacae | | 分子名称: | Ankyrin repeat domain-containing protein, CITRIC ACID, GLYCEROL, ... | | 著者 | Cuthbert, B.J, Jensen, S.J, Goulding, C.W, Hayes, C.S. | | 登録日 | 2024-08-02 | | 公開日 | 2024-08-14 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Advanced glycation end-product (AGE) crosslinking activates a type 6 secretion system phospholipase effector protein
Nature Communications, 2024
|
|
9CX6
 
 | |
9BUV
 
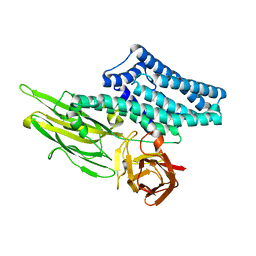 | |
7S4I
 
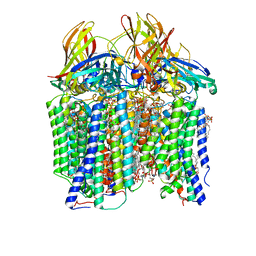 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.26 Angstrom resolution | | 分子名称: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | 著者 | Koo, C.W, Rosenzweig, A.C. | | 登録日 | 2021-09-09 | | 公開日 | 2022-03-30 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.26 Å) | | 主引用文献 | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
8VAX
 
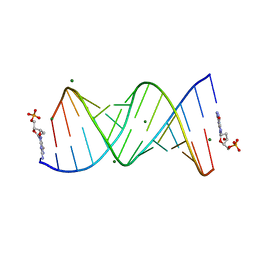 | |
8HCZ
 
 | |
8HDF
 
 | | Full length crystal structure of mycobacterium tuberculosis FadD23 in complex with ANP and PLM | | 分子名称: | Long-chain-fatty-acid--AMP ligase FadD23, PALMITIC ACID, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Yan, M.R, Liu, X, Zhang, W, Rao, Z.H. | | 登録日 | 2022-11-04 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | The Key Roles of Mycobacterium tuberculosis FadD23 C-terminal Domain in Catalytic Mechanisms.
Front Microbiol, 14, 2023
|
|
