6H1S
 
 | | Structure of the BM3 heme domain in complex with fluconazole | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Jeffreys, L.N, Munro, A.W.M, Leys, D. | | Deposit date: | 2018-07-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel insights into P450 BM3 interactions with FDA-approved antifungal azole drugs.
Sci Rep, 9, 2019
|
|
2RFX
 
 | | Crystal Structure of HLA-B*5701, presenting the self peptide, LSSPVTKSF | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Archbold, J.K, Macdonald, W.A, Rossjohn, J, McCluskey, J. | | Deposit date: | 2007-10-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human leukocyte antigen class I-restricted activation of CD8+ T cells provides the immunogenetic basis of a systemic drug hypersensitivity
Immunity, 28, 2008
|
|
6XQQ
 
 | | Structure of human D462-E4 TCR | | Descriptor: | GLYCEROL, TRAV12-2 alpha chain, TRBV29-1 | | Authors: | Awad, W, Rossjohn, J. | | Deposit date: | 2020-07-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Atypical TRAV1-2 - T cell receptor recognition of the antigen-presenting molecule MR1.
J.Biol.Chem., 295, 2020
|
|
6XT6
 
 | | pro-concanavalin A: Precursor of circularly permuted concanavalin A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Concanavalin-A, ... | | Authors: | Nonis, S.G, Haywood, J, Schmidberger, J.W, Bond, C.S. | | Deposit date: | 2020-07-17 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Structural and biochemical analyses of concanavalin A circular permutation by jack bean asparaginyl endopeptidase.
Plant Cell, 33, 2021
|
|
2ROW
 
 | | The C1 domain of ROCK II | | Descriptor: | Rho-associated protein kinase 2, ZINC ION | | Authors: | Wen, W, Zhang, M. | | Deposit date: | 2008-04-25 | | Release date: | 2008-06-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The C1 domain of ROCK II
To be Published
|
|
6U9D
 
 | |
8ZQF
 
 | |
7AB1
 
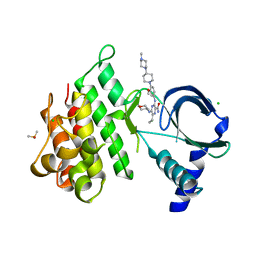 | | Crystal structure of MerTK kinase domain in complex with Gilteritinib | | Descriptor: | 6-ethyl-3-[[3-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]amino]-5-(oxan-4-ylamino)pyrazine-2-carboxamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Overman, R.C, Rawlins, P.B, Truman, C, Underwood, E, Warwicker, J, Winter-Holt, J. | | Deposit date: | 2020-09-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding.
Biochem.J., 477, 2020
|
|
5IMI
 
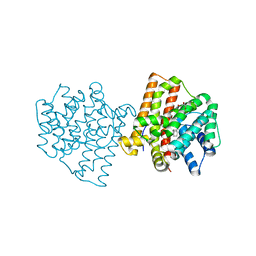 | | Crystal structure of S303A Aspergillus terreus aristolochene synthase complexed with (1S,8S,9aR)-1,9a-dimethyl-8-(prop-1-en-2-yl)decahydroquinolizin-5-ium | | Descriptor: | (1S,5S,8S,9aR)-1,9a-dimethyl-8-(prop-1-en-2-yl)octahydro-2H-quinolizinium, Aristolochene synthase, GLYCEROL, ... | | Authors: | Chen, M, Christianson, D.W. | | Deposit date: | 2016-03-06 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.462 Å) | | Cite: | Probing the Role of Active Site Water in the Sesquiterpene Cyclization Reaction Catalyzed by Aristolochene Synthase.
Biochemistry, 55, 2016
|
|
7AAX
 
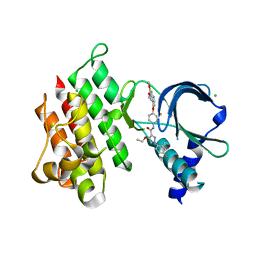 | | Crystal structure of MerTK kinase domain in complex with LDC1267 | | Descriptor: | CHLORIDE ION, Tyrosine-protein kinase Mer, ~{N}-[4-(6,7-dimethoxyquinolin-4-yl)oxy-3-fluoranyl-phenyl]-4-ethoxy-1-(4-fluoranyl-2-methyl-phenyl)pyrazole-3-carboxamide | | Authors: | Schimpl, M, Pflug, A, McCoull, W, Nissink, J.W.M, Overman, R.C, Rawlins, P.B, Truman, C, Underwood, E, Warwicker, J, Winter-Holt, J. | | Deposit date: | 2020-09-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding.
Biochem.J., 477, 2020
|
|
7AB0
 
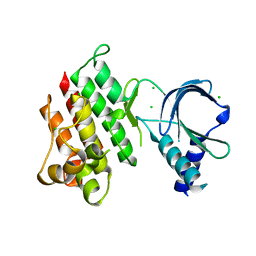 | | Apo crystal structure of the MerTK kinase domain | | Descriptor: | CHLORIDE ION, Tyrosine-protein kinase Mer | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Overman, R.C, Rawlins, P.B, Truman, C, Underwood, E, Warwicker, J, Winter-Holt, J. | | Deposit date: | 2020-09-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding.
Biochem.J., 477, 2020
|
|
4Z3X
 
 | | Active site complex BamBC of Benzoyl Coenzyme A reductase in complex with 1-Monoenoyl-CoA | | Descriptor: | 1,5 Dienoyl-CoA, Benzoyl-CoA reductase, putative, ... | | Authors: | Weinert, T, Kung, J.W, Weidenweber, S, Huwiler, S.G, Boll, M, Ermler, U. | | Deposit date: | 2015-04-01 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of enzymatic benzene ring reduction.
Nat.Chem.Biol., 11, 2015
|
|
2ROH
 
 | | The DNA binding domain of RTBP1 | | Descriptor: | Telomere binding protein-1 | | Authors: | Lee, W, Ko, S. | | Deposit date: | 2008-03-22 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of rice telomere binding protein RTBP1
Biochemistry, 48, 2009
|
|
2TDT
 
 | |
6XB9
 
 | | Crystal structure of Azotobacter vinelandii 3-mercaptopropionic acid dioxygenase in complex with 3-hydroxypropionic acid | | Descriptor: | 3-HYDROXY-PROPANOIC ACID, CHLORIDE ION, Cysteine dioxygenase type I protein, ... | | Authors: | Kiser, P.D, Khadka, N, Shi, W, Pierce, B.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of 3-mercaptopropionic acid dioxygenase with a substrate analog reveals bidentate substrate binding at the iron center.
J.Biol.Chem., 296, 2021
|
|
1R54
 
 | | Crystal structure of the catalytic domain of human ADAM33 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADAM 33, CALCIUM ION, ... | | Authors: | Orth, P, Reicher, P, Wang, W, Prosise, W.W, Yarosh-Tomaine, T, Hammond, G, Xiao, L, Mirza, U.A, Zou, J, Strickland, C, Taremi, S.S. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-12 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structre of the catalytic domain of human ADAM33
J.Mol.Biol., 335, 2004
|
|
6QCH
 
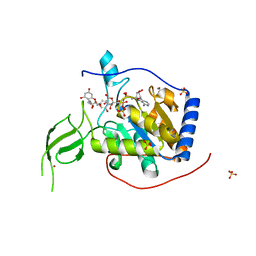 | |
6XL9
 
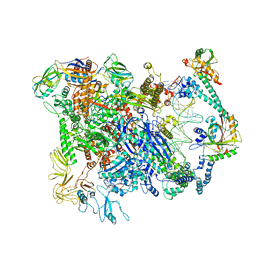 | | Cryo-EM structure of EcmrR-RNAP-promoter initial transcribing complex with 3-nt RNA transcript (EcmrR-RPitc-3nt) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Yang, Y, Liu, C, Shi, W, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XLA
 
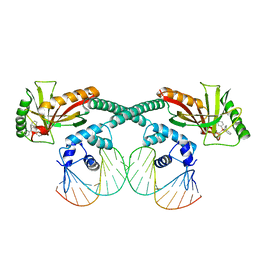 | | Cryo-EM structure of EcmrR-DNA complex in EcmrR-RPitc-3nt | | Descriptor: | MerR family transcriptional regulator EcmrR, TETRAPHENYLANTIMONIUM ION, synthetic non-template strand DNA (54-MER), ... | | Authors: | Yang, Y, Liu, C, Shi, W, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XH7
 
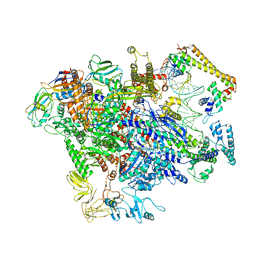 | | CueR-TAC without RNA | | Descriptor: | COPPER (II) ION, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Shi, W, Yang, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of copper-efflux-regulator-dependent transcription activation.
Iscience, 24, 2021
|
|
8D56
 
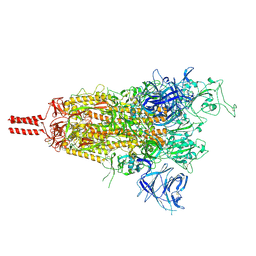 | | One RBD-up state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6XH8
 
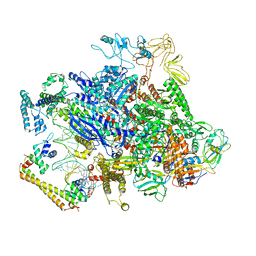 | | CueR-transcription activation complex with RNA transcript | | Descriptor: | COPPER (II) ION, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Shi, W, Yang, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of copper-efflux-regulator-dependent transcription activation.
Iscience, 24, 2021
|
|
8D55
 
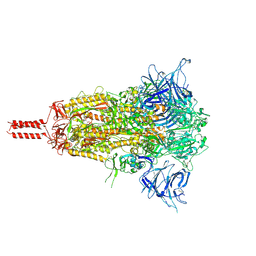 | | Closed state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5FJ8
 
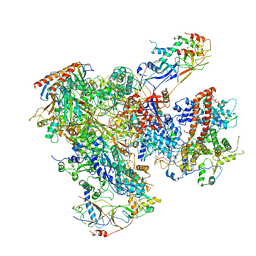 | | Cryo-EM structure of yeast RNA polymerase III elongation complex at 3. 9 A | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC1, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC10, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC2, ... | | Authors: | Hoffmann, N.A, Jakobi, A.J, Moreno-Morcillo, M, Glatt, S, Kosinski, J, Hagen, W.J, Sachse, C, Muller, C.W. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular Structures of Unbound and Transcribing RNA Polymerase III.
Nature, 528, 2015
|
|
1RDL
 
 | | MANNOSE-BINDING PROTEIN, SUBTILISIN DIGEST FRAGMENT COMPLEX WITH ALPHA-METHYL-D-MANNOPYRANOSIDE (0.2 M) | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN-C, ... | | Authors: | Ng, K.K.-S, Drickamer, K, Weis, W.I. | | Deposit date: | 1995-09-05 | | Release date: | 1996-03-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of monosaccharide recognition by rat liver mannose-binding protein.
J.Biol.Chem., 271, 1996
|
|
