7B3K
 
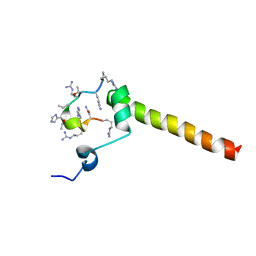 | | Dynamic complex between all-D-enantiomeric peptide D3 with L723P mutant of amyloid precursor protein (APP) 672-726 fragment (amyloid beta 1-55) | | Descriptor: | D3 all D-enantimeric peptide, Isoform L-APP677 of Amyloid-beta precursor protein | | Authors: | Bocharov, E.V, Volynsky, P.E, Okhrimenko, I.S, Urban, A.S. | | Deposit date: | 2020-12-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | All - d - Enantiomeric Peptide D3 Designed for Alzheimer's Disease Treatment Dynamically Interacts with Membrane-Bound Amyloid-beta Precursors.
J.Med.Chem., 64, 2021
|
|
7B3J
 
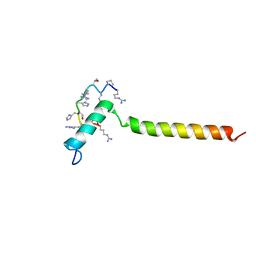 | | Dynamic complex between all-D-enantiomeric peptide D3 with wild-type amyloid precursor protein 672-726 fragment (amyloid beta 1-55) | | Descriptor: | D3 all D-enantimeric peptide, Isoform L-APP677 of Amyloid-beta precursor protein | | Authors: | Bocharov, E.V, Volynsky, P.E, Okhrimenko, I.S, Urban, A.S. | | Deposit date: | 2020-12-01 | | Release date: | 2021-01-13 | | Last modified: | 2021-12-08 | | Method: | SOLUTION NMR | | Cite: | All - d - Enantiomeric Peptide D3 Designed for Alzheimer's Disease Treatment Dynamically Interacts with Membrane-Bound Amyloid-beta Precursors.
J.Med.Chem., 64, 2021
|
|
2KPF
 
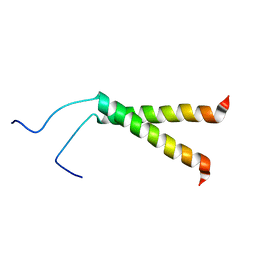 | | Spatial structure of the dimeric transmembrane domain of glycophorin A in bicelles soluton | | Descriptor: | Glycophorin-A | | Authors: | Mineev, K.S, Bocharov, E.V, Goncharuk, M.V, Arseniev, A.S, Volynsky, P.E, Efremov, R.G. | | Deposit date: | 2009-10-13 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Dimeric structure of the transmembrane domain of glycophorin a in lipidic and detergent environments.
Acta Naturae, 3, 2011
|
|
2PCO
 
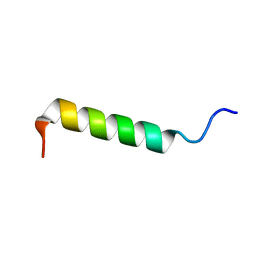 | | Spatial Structure and Membrane Permeabilization for Latarcin-1, a Spider Antimicrobial Peptide | | Descriptor: | Latarcin-1 | | Authors: | Dubovskii, P.V, Volynsky, P.E, Polyansky, A.A, Chupin, V.V, Efremov, R.G, Arseniev, A.S. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure/hydrophobicity of latarcins specifies their mode of membrane activity.
Biochemistry, 47, 2008
|
|
2G9P
 
 | | NMR structure of a novel antimicrobial peptide, latarcin 2a, from spider (Lachesana tarabaevi) venom | | Descriptor: | antimicrobial peptide Latarcin 2a | | Authors: | Dubovskii, P.V, Volynsky, P.E, Polyansky, A.A, Chupin, V.V, Efremov, R.G, Arseniev, A.S. | | Deposit date: | 2006-03-07 | | Release date: | 2006-09-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Spatial structure and activity mechanism of a novel spider antimicrobial peptide.
Biochemistry, 45, 2006
|
|
2J5D
 
 | | NMR structure of BNIP3 transmembrane domain in lipid bicelles | | Descriptor: | BCL2/ADENOVIRUS E1B 19 KDA PROTEIN-INTERACTING PROTEIN 3 | | Authors: | Bocharov, E.V, Pustovalova, Y.E, Volynsky, P.E, Maslennikov, I.V, Goncharuk, M.V, Ermolyuk, Y.S, Arseniev, A.S. | | Deposit date: | 2006-09-14 | | Release date: | 2007-04-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unique dimeric structure of BNip3 transmembrane domain suggests membrane permeabilization as a cell death trigger.
J. Biol. Chem., 282, 2007
|
|
2K9Y
 
 | |
2KPE
 
 | | Refined structure of Glycophorin A transmembrane segment dimer in DPC micelles | | Descriptor: | Glycophorin-A | | Authors: | Mineev, K.S, Bocharov, E.V, Goncharuk, M.V, Arseniev, A.S, Volynsky, P.E, Efremov, R.G. | | Deposit date: | 2009-10-13 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Dimeric structure of the transmembrane domain of glycophorin a in lipidic and detergent environments.
Acta Naturae, 3, 2011
|
|
5NQ4
 
 | | Cytotoxin-1 in DPC-micelle | | Descriptor: | Cytotoxin 1 | | Authors: | Dubovskii, P.V, Dubinnyi, M.A, Volynsky, P.E, Pustovalova, Y.E, Konshina, A.G, Utkin, Y.N, Efremov, R.G, Arseniev, A.S. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-13 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Impact of membrane partitioning on the spatial structure of an S-type cobra cytotoxin.
J. Biomol. Struct. Dyn., 36, 2018
|
|
