7P8W
 
 | | Human erythrocyte catalase cryoEM | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chen, S, Li, J, Vinothkumar, K.R, Henderson, R. | | Deposit date: | 2021-07-23 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Interaction of human erythrocyte catalase with air-water interface in cryoEM.
Microscopy (Oxf), 71, 2022
|
|
7P2L
 
 | | thermostabilised 7TM domain of human mGlu5 receptor bound to photoswitchable ligand alloswitch-1 | | Descriptor: | 2-chloranyl-~{N}-[2-methoxy-4-[(~{E})-pyridin-2-yldiazenyl]phenyl]benzamide, Metabotropic glutamate receptor 5,Endolysin,Metabotropic glutamate receptor 5 | | Authors: | Huang, C.Y, Vinothkumar, K.R, Lebon, G. | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Agonists and allosteric modulators promote signaling from different metabotropic glutamate receptor 5 conformations.
Cell Rep, 36, 2021
|
|
6LVB
 
 | | Structure of Dimethylformamidase, tetramer | | Descriptor: | FE (III) ION, N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
2IWV
 
 | | Structure of the monomeric outer membrane porin OmpG in the open and closed conformation | | Descriptor: | CALCIUM ION, LAURYL DIMETHYLAMINE-N-OXIDE, OUTER MEMBRANE PROTEIN G, ... | | Authors: | Yildiz, O, Vinothkumar, K.R, Goswami, P, Kuehlbrandt, W. | | Deposit date: | 2006-07-04 | | Release date: | 2006-08-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Monomeric Outer-Membrane Porin Ompg in the Open and Closed Conformation.
Embo J., 25, 2006
|
|
2IWW
 
 | | Structure of the monomeric outer membrane porin OmpG in the open and closed conformation | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, OUTER MEMBRANE PROTEIN G, beta-D-glucopyranose, ... | | Authors: | Yildiz, O, Vinothkumar, K.R, Goswami, P, Kuehlbrandt, W. | | Deposit date: | 2006-07-05 | | Release date: | 2006-08-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Monomeric Outer-Membrane Porin Ompg in the Open and Closed Conformation.
Embo J., 25, 2006
|
|
6LVE
 
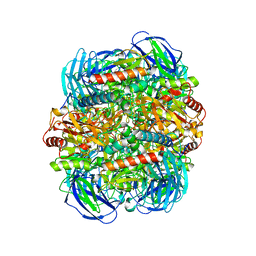 | | Structure of Dimethylformamidase, tetramer, E521A mutant | | Descriptor: | N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6LVC
 
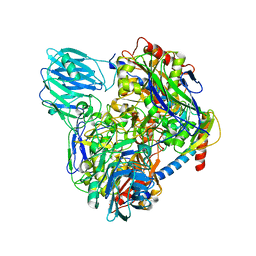 | | Structure of Dimethylformamidase, dimer | | Descriptor: | FE (III) ION, N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6LVD
 
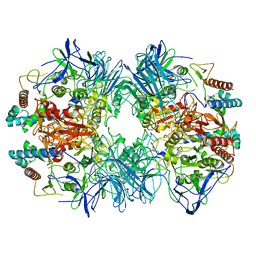 | | Structure of Dimethylformamidase, tetramer, Y440A mutant | | Descriptor: | N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8WV5
 
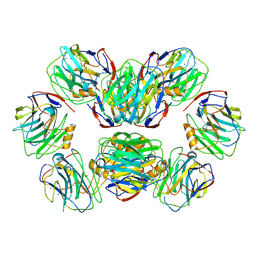 | | C-reactive protein, decamer | | Descriptor: | C-reactive protein(1-205), CALCIUM ION | | Authors: | Yadav, S, Vinothkumar, K.R. | | Deposit date: | 2023-10-23 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Factors affecting macromolecule orientations in thin films formed in cryo-EM.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8WV6
 
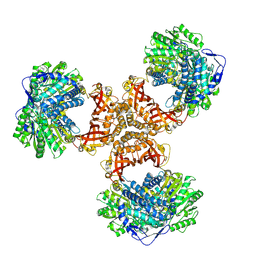 | | PaaZ, bifunctional enzyme | | Descriptor: | Bifunctional protein PaaZ | | Authors: | Yadav, S, Vinothkumar, K.R. | | Deposit date: | 2023-10-23 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Factors affecting macromolecule orientations in thin films formed in cryo-EM.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8WV4
 
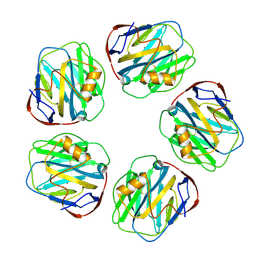 | | C-reactive protein, pentamer | | Descriptor: | C-reactive protein(1-205), CALCIUM ION | | Authors: | Yadav, S, Vinothkumar, K.R. | | Deposit date: | 2023-10-23 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Factors affecting macromolecule orientations in thin films formed in cryo-EM.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8WZM
 
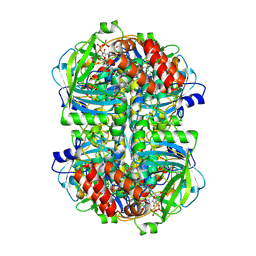 | |
8WZK
 
 | | Human erythrocyte catalase | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yadav, S, Vinothkumar, K.R. | | Deposit date: | 2023-11-01 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Factors affecting macromolecule orientations in thin films formed in cryo-EM.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8WZH
 
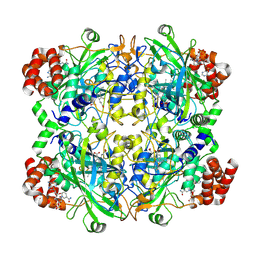 | |
8WZJ
 
 | | Human erythrocyte catalase | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yadav, S, Vinothkumar, K.R. | | Deposit date: | 2023-11-01 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Factors affecting macromolecule orientations in thin films formed in cryo-EM.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8WZI
 
 | | One RBD up state of Spike glycoprotein, SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yadav, S, Vinothkumar, K.R. | | Deposit date: | 2023-11-01 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Factors affecting macromolecule orientations in thin films formed in cryo-EM.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6JQN
 
 | | Structure of PaaZ, a bifunctional enzyme in complex with NADP+ and OCoA | | Descriptor: | Bifunctional protein PaaZ, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OCTANOYL-COENZYME A | | Authors: | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6JQO
 
 | | Structure of PaaZ, a bifunctional enzyme in complex with NADP+ and CCoA | | Descriptor: | Bifunctional protein PaaZ, CROTONYL COENZYME A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6JQL
 
 | | Structure of PaaZ, a bifunctional enzyme | | Descriptor: | Bifunctional protein PaaZ | | Authors: | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6JQM
 
 | | Structure of PaaZ with NADPH | | Descriptor: | Bifunctional protein PaaZ, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
7V2N
 
 | | T.thermophilus 30S ribosome with KsgA, class K2 | | Descriptor: | 16s ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Raina, R, Singh, J, Anand, R, Vinothkumar, K.R. | | Deposit date: | 2021-08-09 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Decoding the Mechanism of Specific RNA Targeting by Ribosomal Methyltransferases.
Acs Chem.Biol., 17, 2022
|
|
7V2O
 
 | | T.thermophilus 30S ribosome with KsgA, class K4 | | Descriptor: | 16s ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Raina, R, Singh, J, Anand, R, Vinothkumar, K.R. | | Deposit date: | 2021-08-09 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Decoding the Mechanism of Specific RNA Targeting by Ribosomal Methyltransferases.
Acs Chem.Biol., 17, 2022
|
|
7V2Q
 
 | | T.thermophilus 30S ribosome with KsgA, class K6 | | Descriptor: | 16s ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Raina, R, Singh, J, Anand, R, Vinothkumar, K.R. | | Deposit date: | 2021-08-09 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Decoding the Mechanism of Specific RNA Targeting by Ribosomal Methyltransferases.
Acs Chem.Biol., 17, 2022
|
|
7V2M
 
 | | T.thermophilus 30S ribosome with KsgA, class K1k4 | | Descriptor: | 16s ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Raina, R, Singh, J, Anand, R, Vinothkumar, K.R. | | Deposit date: | 2021-08-09 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Decoding the Mechanism of Specific RNA Targeting by Ribosomal Methyltransferases.
Acs Chem.Biol., 17, 2022
|
|
7V2P
 
 | | T.thermophilus 30S ribosome with KsgA, class K5 | | Descriptor: | 16s ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Raina, R, Singh, J, Anand, R, Vinothkumar, K.R. | | Deposit date: | 2021-08-09 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Decoding the Mechanism of Specific RNA Targeting by Ribosomal Methyltransferases.
Acs Chem.Biol., 17, 2022
|
|
