2KUY
 
 | | Structure of Glycocin F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prebacteriocin glycocin F | | Authors: | Venugopal, H, Edwards, P, Schwalbe, M, Claridge, J, Stepper, J, Patchett, M, Loo, T, Libich, D, Norris, G, Pascal, S. | | Deposit date: | 2010-03-01 | | Release date: | 2011-05-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural, dynamic, and chemical characterization of a novel s-glycosylated bacteriocin.
Biochemistry, 50, 2011
|
|
8DQV
 
 | | The 1.52 angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - catalytic dimer (Huc2S2L) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-07-20 | | Release date: | 2023-01-04 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (1.52 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
7LLY
 
 | | Oxyntomodulin-bound Glucagon-Like Peptide-1 (GLP-1) Receptor in complex with Gs protein | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wootten, D, Sexton, P.M, Belousoff, M.J, Danev, R, Zhang, X, Khoshouei, M, Venugopal, H. | | Deposit date: | 2021-02-04 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dynamics of GLP-1R peptide agonist engagement are correlated with kinetics of G protein activation.
Nat Commun, 13, 2022
|
|
7LLL
 
 | | Exendin-4-bound Glucagon-Like Peptide-1 (GLP-1) Receptor in complex with Gs protein | | Descriptor: | Exendin-4, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wootten, D, Sexton, P.M, Belousoff, M.J, Danev, R, Zhang, X, Khoshouei, M, Venugopal, H. | | Deposit date: | 2021-02-04 | | Release date: | 2022-01-12 | | Last modified: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Dynamics of GLP-1R peptide agonist engagement are correlated with kinetics of G protein activation.
Nat Commun, 13, 2022
|
|
9C4O
 
 | |
8GHD
 
 | | The structure of h12-LOX in hexameric form bound to inhibitor ML355 and arachidonic acid | | Descriptor: | ARACHIDONIC ACID, FE (II) ION, N-(1,3-benzothiazol-2-yl)-4-{[(2-hydroxy-3-methoxyphenyl)methyl]amino}benzene-1-sulfonamide, ... | | Authors: | Black, K.A, Mobbs, J.I, Venugopal, H, Thal, D.M, Glukhova, A. | | Deposit date: | 2023-03-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM structures of human arachidonate 12S-lipoxygenase bound to endogenous and exogenous inhibitors.
Blood, 142, 2023
|
|
8GHE
 
 | | The structure of h12-LOX in tetrameric form bound to endogenous inhibitor oleoyl-CoA | | Descriptor: | FE (II) ION, Polyunsaturated fatty acid lipoxygenase ALOX12, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name) | | Authors: | Black, K.A, Mobbs, J.I, Venugopal, H, Thal, D.M, Glukhova, A. | | Deposit date: | 2023-03-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.05 Å) | | Cite: | Cryo-EM structures of human arachidonate 12S-lipoxygenase bound to endogenous and exogenous inhibitors.
Blood, 142, 2023
|
|
8GHC
 
 | | The structure of h12-LOX in dimeric form | | Descriptor: | FE (II) ION, Polyunsaturated fatty acid lipoxygenase ALOX12 | | Authors: | Black, K.A, Mobbs, J.I, Venugopal, H, Thal, D.M, Glukhova, A. | | Deposit date: | 2023-03-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM structures of human arachidonate 12S-lipoxygenase bound to endogenous and exogenous inhibitors.
Blood, 142, 2023
|
|
8GHB
 
 | | The structure of h12-LOX in monomeric form | | Descriptor: | FE (II) ION, Polyunsaturated fatty acid lipoxygenase ALOX12 | | Authors: | Black, K.A, Mobbs, J.I, Venugopal, H, Thal, D.M, Glukhova, A. | | Deposit date: | 2023-03-09 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Cryo-EM structures of human arachidonate 12S-lipoxygenase bound to endogenous and exogenous inhibitors.
Blood, 142, 2023
|
|
6DDD
 
 | | Structure of the 50S ribosomal subunit from Methicillin Resistant Staphylococcus aureus in complex with the oxazolidinone antibiotic LZD-5 | | Descriptor: | 2,2-dichloro-N-({(5S)-3-[3-fluoro-4-(morpholin-4-yl)phenyl]-2-oxo-1,3-oxazolidin-5-yl}methyl)acetamide, 23S rRNA, 50S ribosomal protein L13, ... | | Authors: | Belousoff, M.J, Venugopal, H, Bamert, R.S, Lithgow, T. | | Deposit date: | 2018-05-10 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | cryoEM-Guided Development of Antibiotics for Drug-Resistant Bacteria.
ChemMedChem, 14, 2019
|
|
6DDG
 
 | | Structure of the 50S ribosomal subunit from Methicillin Resistant Staphylococcus aureus in complex with the oxazolidinone antibiotic LZD-6 | | Descriptor: | 2-chloro-N-({(5S)-3-[3-fluoro-4-(morpholin-4-yl)phenyl]-2-oxo-1,3-oxazolidin-5-yl}methyl)acetamide, 23S rRNA, 50S ribosomal protein L13, ... | | Authors: | Belousoff, M.J, Venugopal, H, Bamert, R.S, Lithgow, T. | | Deposit date: | 2018-05-10 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | cryoEM-Guided Development of Antibiotics for Drug-Resistant Bacteria.
ChemMedChem, 14, 2019
|
|
6D9H
 
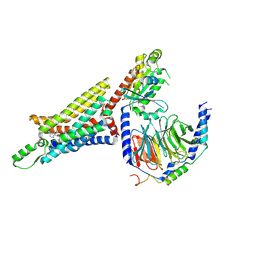 | | Cryo-EM structure of the human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist | | Descriptor: | ADENOSINE, Chimera protein of Muscarinic acetylcholine receptor M4 and Adenosine receptor A1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Draper-Joyce, C.J, Khoshouei, M, Thal, D.M, Liang, Y.-L, Nguyen, A.T.N, Furness, S.G.B, Venugopal, H, Baltos, J, Plitzko, J.M, Danev, R, Baumeister, W, May, L.T, Wootten, D, Sexton, P, Glukhova, A, Christopoulos, A. | | Deposit date: | 2018-04-29 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the adenosine-bound human adenosine A1receptor-Gicomplex.
Nature, 558, 2018
|
|
6XGR
 
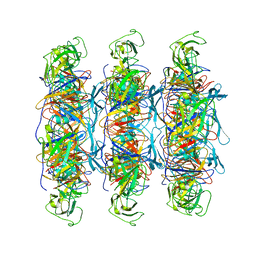 | | YSD1 major tail protein | | Descriptor: | YSD1_22 major tail protein | | Authors: | Hardy, J.M, Dunstan, R, Venugopal, H, Lithgow, T.J, Coulibaly, F.J. | | Deposit date: | 2020-06-17 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The architecture and stabilisation of flagellotropic tailed bacteriophages.
Nat Commun, 11, 2020
|
|
6XGQ
 
 | | YSD1 bacteriophage capsid | | Descriptor: | YSD1_16, YSD1_17 | | Authors: | Hardy, J.M, Dunstan, R, Venugopal, H, Lithgow, T.J, Coulibaly, F.J. | | Deposit date: | 2020-06-17 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The architecture and stabilisation of flagellotropic tailed bacteriophages.
Nat Commun, 11, 2020
|
|
7UUR
 
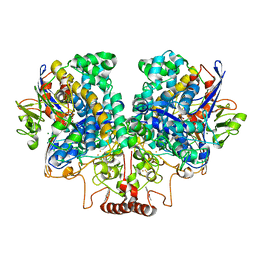 | | The 1.67 Angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - catalytic dimer (Huc2S2L) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, HYDROXIDE ION, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-04 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (1.67 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
7UUS
 
 | | The CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - Full complex focused refinement of stalk | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
7UTD
 
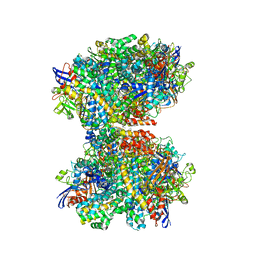 | | The 2.19-angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - Complex minus stalk | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-04-26 | | Release date: | 2023-01-04 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
7KVB
 
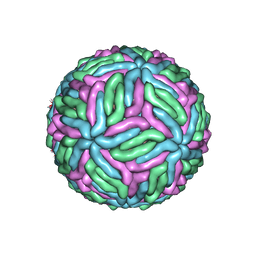 | | Chimeric flavivirus between Binjari virus and Murray Valley encephalitis virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, Matrix protein M | | Authors: | Hardy, J.M, Venugopal, H.V, Newton, N.D, Watterson, D, Coulibaly, F.J. | | Deposit date: | 2020-11-27 | | Release date: | 2020-12-23 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A unified route for flavivirus structures uncovers essential pocket factors conserved across pathogenic viruses.
Nat Commun, 12, 2021
|
|
7KV9
 
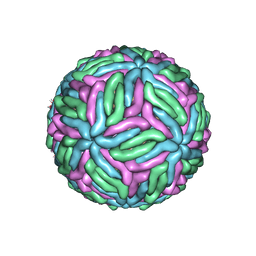 | | Chimeric flavivirus between Binjari virus and West Nile (Kunjin) virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Matrix protein M, envelope protein E | | Authors: | Hardy, J.M, Venugopal, H.V, Newton, N.D, Watterson, D, Coulibaly, F.J. | | Deposit date: | 2020-11-27 | | Release date: | 2020-12-23 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A unified route for flavivirus structures uncovers essential pocket factors conserved across pathogenic viruses.
Nat Commun, 12, 2021
|
|
8UY4
 
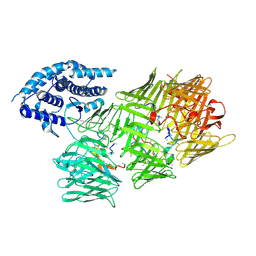 | | Acinetobacter baumannii Tse15 Rhs effector | | Descriptor: | Tse15, Tse15 toxin peptide (polyUNK) | | Authors: | Hayes, B.K, Venugopal, H, McGowan, S. | | Deposit date: | 2023-11-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structure of a Rhs effector clade domain provides mechanistic insights into type VI secretion system toxin delivery.
Nat Commun, 15, 2024
|
|
8UXT
 
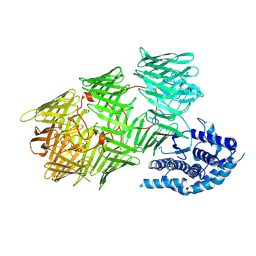 | |
7KV8
 
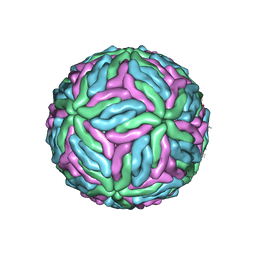 | | Chimeric flavivirus between Binjari virus and Dengue virus serotype-2 | | Descriptor: | (2S,3R,4Z)-3-hydroxy-2-[(9E)-octadec-9-enoylamino]octadec-4-en-1-yl dihydrogen phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Hardy, J.M, Venugopal, H.V, Newton, N.D, Watterson, D, Coulibaly, F.J. | | Deposit date: | 2020-11-27 | | Release date: | 2020-12-23 | | Last modified: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A unified route for flavivirus structures uncovers essential pocket factors conserved across pathogenic viruses.
Nat Commun, 12, 2021
|
|
7KVA
 
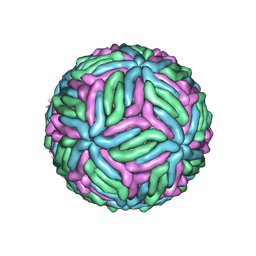 | | Structure of West Nile virus (Kunjin) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, Matrix protein M | | Authors: | Hardy, J.M, Venugopal, H.V, Newton, N.D, Watterson, D, Coulibaly, F.J. | | Deposit date: | 2020-11-27 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A unified route for flavivirus structures uncovers essential pocket factors conserved across pathogenic viruses.
Nat Commun, 12, 2021
|
|
7L30
 
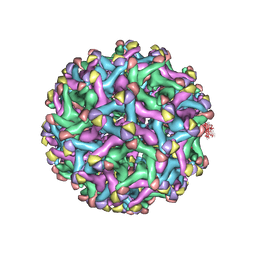 | | Binjari virus (BinJV) | | Descriptor: | Envelope protein E, prM protein | | Authors: | Hardy, J.M, Venugopal, H.V, Newton, N.D, Watterson, D, Coulibaly, F.J. | | Deposit date: | 2020-12-17 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The structure of an infectious immature flavivirus redefines viral architecture and maturation.
Sci Adv, 7, 2021
|
|
