1RY3
 
 | | NMR Solution Structure of the Precursor for Carnobacteriocin B2, an Antimicrobial Peptide from Carnobacterium piscicola | | Descriptor: | Bacteriocin carnobacteriocin B2 | | Authors: | Sprules, T, Kawulka, K.E, Gibbs, A.C, Wishart, D.S, Vederas, J.C. | | Deposit date: | 2003-12-19 | | Release date: | 2004-05-04 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the precursor for carnobacteriocin B2, an antimicrobial peptide from Carnobacterium piscicola.
Eur.J.Biochem., 271, 2004
|
|
1QA7
 
 | | CRYSTAL COMPLEX OF THE 3C PROTEINASE FROM HEPATITIS A VIRUS WITH ITS INHIBITOR AND IMPLICATIONS FOR THE POLYPROTEIN PROCESSING IN HAV | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, HAV 3C PROTEINASE, ... | | Authors: | Bergmann, E.M, Cherney, M.M, Mckendrick, J, Vederas, J.C, James, M.N.G. | | Deposit date: | 1999-04-15 | | Release date: | 1999-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an inhibitor complex of the 3C proteinase from hepatitis A virus (HAV) and implications for the polyprotein processing in HAV.
Virology, 265, 1999
|
|
2GKE
 
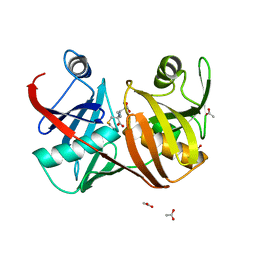 | | Crystal structure of diaminopimelate epimerase in complex with an irreversible inhibitor LL-AziDAP | | Descriptor: | (2S,6S)-2,6-DIAMINO-2-METHYLHEPTANEDIOIC ACID, ACETIC ACID, Diaminopimelate epimerase, ... | | Authors: | Pillai, B, Cherney, M.M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C, James, M.N. | | Deposit date: | 2006-04-01 | | Release date: | 2006-05-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into stereochemical inversion by diaminopimelate epimerase: An antibacterial drug target.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2CXV
 
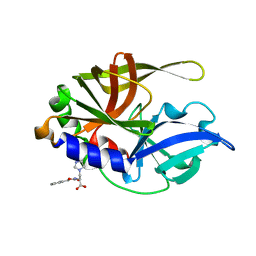 | | Dual Modes of Modification of Hepatitis A Virus 3C Protease by a Serine-Derived betaLactone: Selective Crystallization and High-resolution Structure of the His-102 Adduct | | Descriptor: | N-[(BENZYLOXY)CARBONYL]-L-ALANINE, Probable protein P3C | | Authors: | Yin, J, Bergmann, E.M, Cherney, M.M, Lall, M.S, Jain, R.P, Vederas, J.C, James, M.N.G. | | Deposit date: | 2005-07-01 | | Release date: | 2005-12-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Dual Modes of Modification of Hepatitis A Virus 3C Protease by a Serine-derived beta-Lactone: Selective Crystallization and Formation of a Functional Catalytic Triad in the Active Site
J.MOL.BIOL., 354, 2005
|
|
2GKJ
 
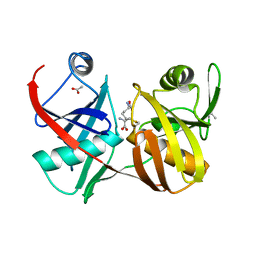 | | Crystal structure of diaminopimelate epimerase in complex with an irreversible inhibitor DL-AZIDAP | | Descriptor: | (2R,6S)-2,6-DIAMINO-2-METHYLHEPTANEDIOIC ACID, ACETIC ACID, Diaminopimelate epimerase | | Authors: | Pillai, B, Cherney, M.M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C, James, M.N. | | Deposit date: | 2006-04-02 | | Release date: | 2006-05-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into stereochemical inversion by diaminopimelate epimerase: An antibacterial drug target.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3DAP
 
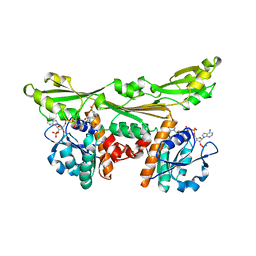 | | C. GLUTAMICUM DAP DEHYDROGENASE IN COMPLEX WITH NADP+ AND THE INHIBITOR 5S-ISOXAZOLINE | | Descriptor: | (2S,5',S)-2-AMINO-3-(3-CARBOXY-2-ISOXAZOLIN-5-YL)PROPANOIC ACID, DIAMINOPIMELIC ACID DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Scapin, G, Cirilli, M, Reddy, S.G, Gao, Y, Vederas, J.C, Blanchard, J.S. | | Deposit date: | 1997-12-29 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate and inhibitor binding sites in Corynebacterium glutamicum diaminopimelate dehydrogenase.
Biochemistry, 37, 1998
|
|
3B6Z
 
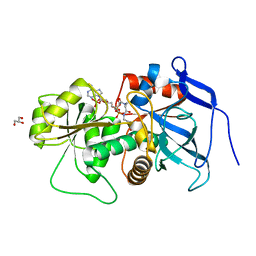 | | Lovastatin polyketide enoyl reductase (LovC) complexed with 2'-phosphoadenosyl isomer of crotonoyl-CoA | | Descriptor: | Enoyl reductase, GLYCEROL, S-{(9R,13R,15S)-17-[(2R,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3-hydroxy-4-(phosphonooxy)tetrahydrofuran-2-yl]-9,13,15-trihydroxy-10,10-dimethyl-13,15-dioxido-4,8-dioxo-12,14,16-trioxa-3,7-diaza-13,15-diphosphaheptadec-1-yl}(2E)-but-2-enethioate | | Authors: | Ames, B.D, Smith, P.T, Ma, S.M, Wong, E.W, Xie, X, Vederas, J.C, Tang, Y, Tsai, S.-C. | | Deposit date: | 2007-10-29 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure and biochemical studies of the trans-acting polyketide enoyl reductase LovC from lovastatin biosynthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3B70
 
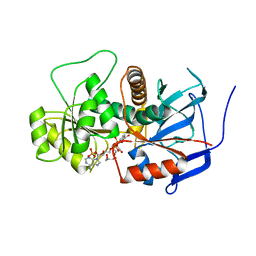 | | Crystal structure of Aspergillus terreus trans-acting lovastatin polyketide enoyl reductase (LovC) with bound NADP | | Descriptor: | Enoyl reductase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ames, B.D, Smith, P.T, Ma, S.M, Wong, E.W, Xie, X, Vederas, J.C, Tang, Y, Tsai, S.-C. | | Deposit date: | 2007-10-29 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure and biochemical studies of the trans-acting polyketide enoyl reductase LovC from lovastatin biosynthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3EI9
 
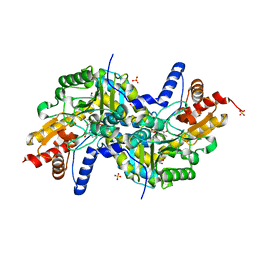 | | Crystal structure of K270N variant of LL-diaminopimelate aminotransferase from Arabidopsis thaliana complexed with L-Glu: External aldimine form | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-glutamic acid, GLYCEROL, LL-diaminopimelate aminotransferase, ... | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
3EI8
 
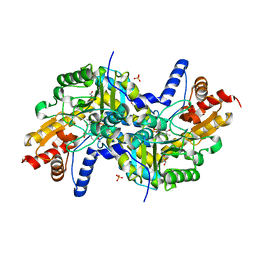 | | Crystal structure of K270N variant of LL-diaminopimelate aminotransferase from Arabidopsis thaliana complexed with LL-DAP: External aldimine form | | Descriptor: | (2S,6S)-2-amino-6-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}heptanedioic acid, GLYCEROL, LL-diaminopimelate aminotransferase, ... | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
3EI6
 
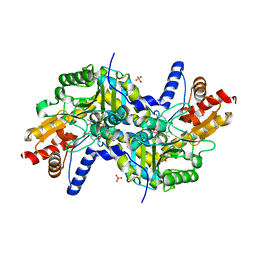 | | Crystal structure of LL-diaminopimelate aminotransferase from Arabidopsis thaliana complexed with PLP-DAP: an external aldimine mimic | | Descriptor: | (2S,6S)-2-amino-6-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]heptanedioic acid, GLYCEROL, LL-diaminopimelate aminotransferase, ... | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
3EIB
 
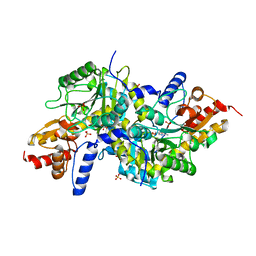 | | Crystal structure of K270N variant of LL-diaminopimelate aminotransferase from Arabidopsis thaliana | | Descriptor: | GLYCEROL, LL-diaminopimelate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
3EJX
 
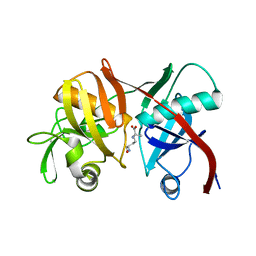 | | Crystal structure of diaminopimelate epimerase from Arabidopsis thaliana in complex with LL-AziDAP | | Descriptor: | (2S,6S)-2,6-DIAMINO-2-METHYLHEPTANEDIOIC ACID, Diaminopimelate epimerase, chloroplastic | | Authors: | Pillai, B, Moorthie, V.A, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-18 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of diaminopimelate epimerase from Arabidopsis thaliana, an amino acid racemase critical for L-lysine biosynthesis.
J.Mol.Biol., 385, 2009
|
|
2LA0
 
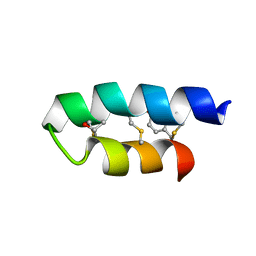 | | Trn- peptide of the two-component bacteriocin Thuricin CD | | Descriptor: | Uncharacterized protein | | Authors: | Sit, C.S, Mckay, R.T, Hill, C, Ross, R.P, Vederas, J.C. | | Deposit date: | 2011-02-27 | | Release date: | 2012-01-11 | | Last modified: | 2018-08-22 | | Method: | SOLUTION NMR | | Cite: | The 3D structure of thuricin CD, a two-component bacteriocin with cysteine sulfur to alpha-carbon cross-links.
J.Am.Chem.Soc., 133, 2011
|
|
2LJT
 
 | | C9L,C14L-LeuA | | Descriptor: | Bacteriocin leucocin-A | | Authors: | Sit, C.S, Lohans, C.T, van Belkum, M.J, Campbell, C.D, Miskolzie, M, Vederas, J.C. | | Deposit date: | 2011-09-23 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Substitution of a Conserved Disulfide in the Type IIa Bacteriocin, Leucocin A, with L-Leucine and L-Serine Residues: Effects on Activity and Three-Dimensional Structure.
Chembiochem, 13, 2012
|
|
2LJQ
 
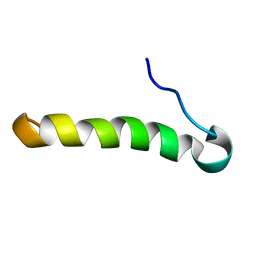 | | (C9S, C14S)-leucocin A | | Descriptor: | Bacteriocin leucocin-A | | Authors: | Sit, C.S, Lohans, C.T, van Belkum, M.J, Campbell, C.D, Miskolzie, M, Vederas, J.C. | | Deposit date: | 2011-09-22 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Substitution of a Conserved Disulfide in the Type IIa Bacteriocin, Leucocin A, with L-Leucine and L-Serine Residues: Effects on Activity and Three-Dimensional Structure.
Chembiochem, 13, 2012
|
|
2M60
 
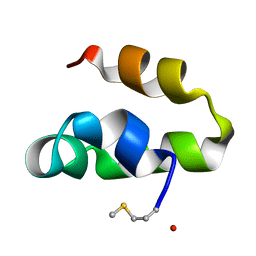 | | Enterocin 7B | | Descriptor: | Enterocin JSB | | Authors: | Lohans, C.T, Towle, K.M, Miskolzie, M, McKay, R.T, van Belkum, M.J, McMullen, L.M, Vederas, J.C. | | Deposit date: | 2013-03-18 | | Release date: | 2013-06-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the Linear Leaderless Bacteriocins Enterocin 7A and 7B Resemble Carnocyclin A, a Circular Antimicrobial Peptide
Biochemistry, 52, 2013
|
|
2M5Z
 
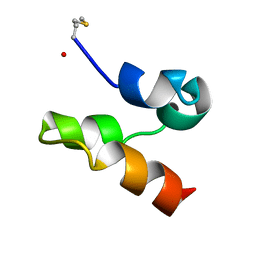 | | Enterocin 7A | | Descriptor: | Enterocin JSA | | Authors: | Lohans, C.T, Towle, K.M, Miskolzie, M, McKay, R.T, van Belkum, M.J, McMullen, L.M, Vederas, J.C. | | Deposit date: | 2013-03-16 | | Release date: | 2013-06-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the Linear Leaderless Bacteriocins Enterocin 7A and 7B Resemble Carnocyclin A, a Circular Antimicrobial Peptide
Biochemistry, 52, 2013
|
|
2K19
 
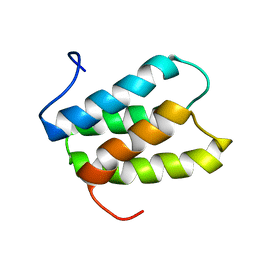 | | NMR solution structure of PisI | | Descriptor: | Putative piscicolin 126 immunity protein | | Authors: | Martin-Visscher, L.A, Sprules, T, Gursky, L.J, Vederas, J.C. | | Deposit date: | 2008-02-25 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of PisI, a group B immunity protein that provides protection against the type IIa bacteriocin piscicolin 126, PisA.
Biochemistry, 47, 2008
|
|
2N4K
 
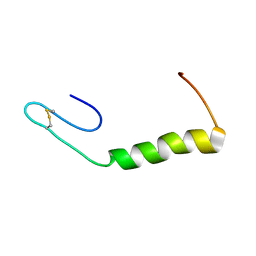 | | Solution Structure of Enterocin HF, an Antilisterial Bacteriocin Produced by Enterococcus faecium M3K31 | | Descriptor: | Enterocin-HF | | Authors: | Arbulu, S, Lohans, C.T, van Belkum, M.J, Cintas, L.M, Herranz, C, Vederas, J.C, Hernandez, P.E. | | Deposit date: | 2015-06-21 | | Release date: | 2015-12-02 | | Last modified: | 2016-01-06 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Enterocin HF, an Antilisterial Bacteriocin Produced by Enterococcus faecium M3K31.
J.Agric.Food Chem., 63, 2015
|
|
2N5W
 
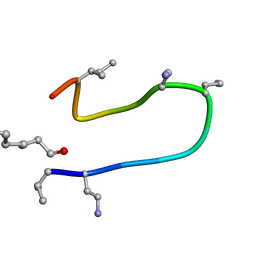 | | The NMR solution structure of octyl-tridecaptin A1 in DPC micelles | | Descriptor: | Octyl-tridecaptin A1 | | Authors: | Cochrane, S.A, Findlay, B, Bakhtiary, A, Acedo, J.Z, Rodriguez-Lopez, E.M, Vederas, J.C. | | Deposit date: | 2015-08-01 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Antimicrobial lipopeptide tridecaptin A1 selectively binds to Gram-negative lipid II.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2LBZ
 
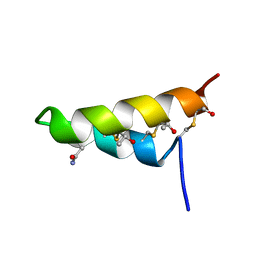 | | Thurincin H | | Descriptor: | Thuricin17 | | Authors: | Sit, C.S, van Belkum, M.J, Mckay, R.T, Worobo, R.W, Vederas, J.C. | | Deposit date: | 2011-04-10 | | Release date: | 2012-01-18 | | Last modified: | 2018-08-22 | | Method: | SOLUTION NMR | | Cite: | The 3D solution structure of thurincin H, a bacteriocin with four sulfur to alpha-carbon crosslinks.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
2L9X
 
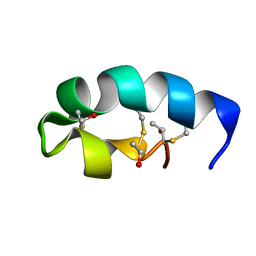 | | Trn- peptide of the two-component bacteriocin Thuricin CD | | Descriptor: | Uncharacterized protein | | Authors: | Sit, C.S, Mckay, R.T, Hill, C, Ross, R.P, Vederas, J.C. | | Deposit date: | 2011-02-25 | | Release date: | 2012-01-11 | | Last modified: | 2018-08-22 | | Method: | SOLUTION NMR | | Cite: | The 3D structure of thuricin CD, a two-component bacteriocin with cysteine sulfur to alpha-carbon cross-links.
J.Am.Chem.Soc., 133, 2011
|
|
2N5Y
 
 | | Solution NMR structure of octyl-tridecaptin A1 in DPC micelles containing Gram-negative lipid II | | Descriptor: | Octyl-tridecaptin A1 | | Authors: | Cochrane, S.A, Findlay, B, Bakhtiary, A, Rodriguez-Lopez, E.M, Vederas, J.C. | | Deposit date: | 2015-08-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Antimicrobial lipopeptide tridecaptin A1 selectively binds to Gram-negative lipid II.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2N8O
 
 | | NMR Solution Structure of Aureocin A53 | | Descriptor: | Bacteriocin aureocin A53 | | Authors: | Acedo, J.Z, van Belkum, M.J, Lohans, C.T, Towle, K.M, Miskolzie, M, Vederas, J.C. | | Deposit date: | 2015-10-22 | | Release date: | 2016-02-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structures of Lacticin Q and Aureocin A53 Reveal a Structural Motif Conserved among Leaderless Bacteriocins with Broad-Spectrum Activity.
Biochemistry, 55, 2016
|
|
