4S2Y
 
 | | Structure of E. coli RppH bound to RNA and three magnesium ions | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, RNA (5'-R(*(APC)*GP*U)-3'), ... | | Authors: | Vasilyev, N, Serganov, A. | | Deposit date: | 2015-01-23 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of RNA Complexes with the Escherichia coli RNA Pyrophosphohydrolase RppH Unveil the Basis for Specific 5'-End-dependent mRNA Decay.
J.Biol.Chem., 290, 2015
|
|
4S2V
 
 | | E. coli RppH structure, KI soak | | Descriptor: | ACETATE ION, CALCIUM ION, IODIDE ION, ... | | Authors: | Vasilyev, N, Serganov, A. | | Deposit date: | 2015-01-23 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of RNA Complexes with the Escherichia coli RNA Pyrophosphohydrolase RppH Unveil the Basis for Specific 5'-End-dependent mRNA Decay.
J.Biol.Chem., 290, 2015
|
|
5DE5
 
 | | Crystal structure of the complex between human FMRP RGG motif and G-quadruplex RNA. | | Descriptor: | Fragile X mental retardation protein 1, POTASSIUM ION, sc1 | | Authors: | Vasilyev, N, Polonskaia, A, Darnell, J.C, Darnell, R.B, Patel, D.J, Serganov, A. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.0011 Å) | | Cite: | Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5DEA
 
 | | Crystal structure of the complex between human FMRP RGG motif and G-quadruplex RNA, cesium bound form. | | Descriptor: | CESIUM ION, Fragile X mental retardation protein 1, POTASSIUM ION, ... | | Authors: | Vasilyev, N, Polonskaia, A, Darnell, J.C, Darnell, R.B, Patel, D.J, Serganov, A. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7973 Å) | | Cite: | Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5DE8
 
 | | Crystal structure of the complex between human FMRP RGG motif and G-quadruplex RNA, iridium hexammine bound form. | | Descriptor: | Fragile X mental retardation protein 1, IRIDIUM HEXAMMINE ION, POTASSIUM ION, ... | | Authors: | Vasilyev, N, Polonskaia, A, Darnell, J.C, Darnell, R.B, Patel, D.J, Serganov, A. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1003 Å) | | Cite: | Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4S2W
 
 | | Structure of E. coli RppH bound to sulfate ions | | Descriptor: | RNA pyrophosphohydrolase, SULFATE ION | | Authors: | Vasilyev, N, Serganov, A. | | Deposit date: | 2015-01-23 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Structures of RNA Complexes with the Escherichia coli RNA Pyrophosphohydrolase RppH Unveil the Basis for Specific 5'-End-dependent mRNA Decay.
J.Biol.Chem., 290, 2015
|
|
4S2X
 
 | | Structure of E. coli RppH bound to RNA and two magnesium ions | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(APC)*GP*U)-3'), RNA pyrophosphohydrolase, ... | | Authors: | Vasilyev, N, Serganov, A. | | Deposit date: | 2015-01-23 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of RNA Complexes with the Escherichia coli RNA Pyrophosphohydrolase RppH Unveil the Basis for Specific 5'-End-dependent mRNA Decay.
J.Biol.Chem., 290, 2015
|
|
7SP3
 
 | | E. coli RppH bound to Ap4A | | Descriptor: | BIS(ADENOSINE)-5'-TETRAPHOSPHATE, CHLORIDE ION, FLUORIDE ION, ... | | Authors: | Serganov, A.A, Vasilyev, N, Nuthanakanti, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A distinct RNA recognition mechanism governs Np 4 decapping by RppH.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6O96
 
 | | Dot1L bound to the H2BK120 Ubiquitinated nucleosome | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Valencia-Sanchez, M.I, De Ioannes, P.E, Miao, W, Vasilyev, N, Chen, R, Nudler, E, Armache, J.-P, Armache, K.-J. | | Deposit date: | 2019-03-13 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Basis of Dot1L Stimulation by Histone H2B Lysine 120 Ubiquitination.
Mol.Cell, 74, 2019
|
|
6VCK
 
 | | Crystal structure of E.coli RppH-DapF in complex with GDP, Mg2+ and F- | | Descriptor: | CHLORIDE ION, Diaminopimelate epimerase, FLUORIDE ION, ... | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6VCP
 
 | | Crystal structure of E.coli RppH in complex with UTP | | Descriptor: | RNA pyrophosphohydrolase, URIDINE 5'-TRIPHOSPHATE | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6VCQ
 
 | | Crystal structure of E.coli RppH in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, RNA pyrophosphohydrolase, SULFATE ION | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6VCN
 
 | | Crystal structure of E.coli RppH in complex with ppcpG | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, RNA pyrophosphohydrolase, SULFATE ION | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6VCM
 
 | | Crystal structure of E.coli RppH-DapF in complex with GTP, Mg2+ and F- | | Descriptor: | CHLORIDE ION, Diaminopimelate epimerase, FLUORIDE ION, ... | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6VCR
 
 | | Crystal structure of E.coli RppH in complex with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, PYROPHOSPHATE, RNA pyrophosphohydrolase, ... | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6VCO
 
 | | Crystal structure of E.coli RppH in complex with ppcpA | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, RNA pyrophosphohydrolase | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6VCL
 
 | | Crystal structure of E.coli RppH-DapF in complex with pppGpp, Mg2+ and F- | | Descriptor: | CHLORIDE ION, Diaminopimelate epimerase, FLUORIDE ION, ... | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6AWD
 
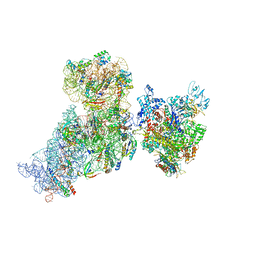 | | Structure of 30S (S1 depleted) ribosomal subunit and RNA polymerase complex | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
6AWB
 
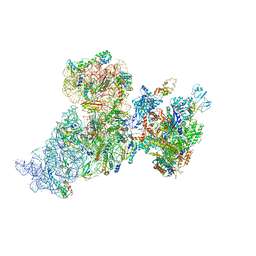 | | Structure of 30S ribosomal subunit and RNA polymerase complex in non-rotated state | | Descriptor: | 16S rRNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
6AWC
 
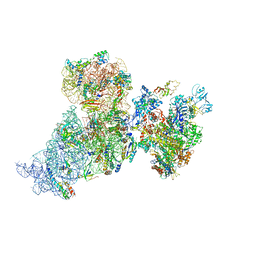 | | Structure of 30S ribosomal subunit and RNA polymerase complex in rotated state | | Descriptor: | 16S rRNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
6QOY
 
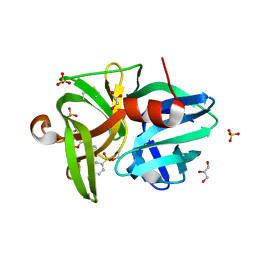 | | Crystal structure of L1 protease Lysobacter sp. XL1 in complex with AEBSF | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, 4-(2-azanylethyl)benzenesulfonic acid, CHLORIDE ION, ... | | Authors: | Gabdulkhakov, A, Tishchenko, S, Kudryakova, I, Afoshin, A, Vasilyeva, N. | | Deposit date: | 2019-02-13 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Serine bacteriolytic protease L1 of Lysobacter sp. XL1 complexed with protease inhibitor AEBSF: features of interaction
Process Biochem, 2019
|
|
8AF1
 
 | | Beta-Lytic Protease from Lysobacter capsici | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Kudryakova, I.V, Afoshin, A.S, Vasilyeva, N.V. | | Deposit date: | 2022-07-15 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural and Functional Characterization of beta-lytic Protease from Lysobacter capsici VKM B-2533 T.
Int J Mol Sci, 23, 2022
|
|
