5LNV
 
 | | Crystal structure of Arabidopsis thaliana Pdx1-I320 complex from multiple crystals | | Descriptor: | (4~{S})-4-azanyl-5-oxidanyl-pent-1-en-3-one, PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3, ... | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
6HJX
 
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) 7'C pore mutant (L238C) in complex with nanobody 72 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cys-loop ligand-gated ion channel, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Spurny, R, Govaerts, C, Evans, G.L, Pardon, E, Steyaert, J, Ulens, C. | | Deposit date: | 2018-09-04 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A lipid site shapes the agonist response of a pentameric ligand-gated ion channel.
Nat.Chem.Biol., 15, 2019
|
|
6HJY
 
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) Delta8 truncation mutant in complex with nanobody 72 | | Descriptor: | Cys-loop ligand-gated ion channel, nanobody 72 | | Authors: | Spurny, R, Govaerts, C, Evans, G.L, Pardon, E, Steyaert, J, Ulens, C. | | Deposit date: | 2018-09-04 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A lipid site shapes the agonist response of a pentameric ligand-gated ion channel.
Nat.Chem.Biol., 15, 2019
|
|
5TIS
 
 | | Room temperature XFEL structure of the native, doubly-illuminated photosystem II complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Fuller, F, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Hussein, R, Zhang, M, Douthit, L, Kubin, M, de Lichtenberg, C, Pham, L.V, Nilsson, H, Cheah, M.H, Shevela, D, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Pastor, E, Weninger, C, Fransson, T, Lassalle, L, Braeuer, P, Aller, P, Docker, P.T, Andi, B, Orville, A.M, Glownia, J.M, Nelson, S, Sikorski, M, Zhu, D, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Watermann, D.G, Evans, G, Wernet, P, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25000381 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
6SSP
 
 | | Structure of the pentameric ligand-gated ion channel ELIC in complex with a NAM nanobody | | Descriptor: | CALCIUM ION, Cys-loop ligand-gated ion channel, NANOBODY 21, ... | | Authors: | Ulens, C, Brams, M, Evans, G.L, Spurny, R, Govaerts, C, Pardon, E, Steyaert, J. | | Deposit date: | 2019-09-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Modulation of the Erwinia ligand-gated ion channel (ELIC) and the 5-HT 3 receptor via a common vestibule site.
Elife, 9, 2020
|
|
6SSI
 
 | | Structure of the pentameric ligand-gated ion channel ELIC in complex with a PAM nanobody | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, CALCIUM ION, ... | | Authors: | Ulens, C, Brams, M, Evans, G.L, Spurny, R, Govaerts, C, Pardon, E, Steyaert, J. | | Deposit date: | 2019-09-07 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Modulation of the Erwinia ligand-gated ion channel (ELIC) and the 5-HT 3 receptor via a common vestibule site.
Elife, 9, 2020
|
|
1NW4
 
 | | Crystal Structure of Plasmodium falciparum Purine Nucleoside Phosphorylase in complex with ImmH and Sulfate | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Shi, W, Ting, L.M, Kicska, G.A, Lewandowicz, A, Tyler, P.C, Evans, G.B, Furneaux, R.H, Kim, K, Almo, S.C, Schramm, V.L. | | Deposit date: | 2003-02-05 | | Release date: | 2004-03-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Plasmodium falciparum Purine Nucleoside Phosphorylase: CRYSTAL STRUCTURES, IMMUCILLIN INHIBITORS, AND DUAL CATALYTIC FUNCTION.
J.Biol.Chem., 279, 2004
|
|
1MHW
 
 | | Design of non-covalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides | | Descriptor: | 4-biphenylacetyl-Cys-(D)Arg-Tyr-N-(2-phenylethyl) amide, Cathepsin L | | Authors: | Chowdhury, S, Sivaraman, J, Wang, J, Devanathan, G, Lachance, P, Qi, H, Menard, R, Lefebvre, J, Konishi, Y, Cygler, M, Sulea, T, Purisima, E.O. | | Deposit date: | 2002-08-21 | | Release date: | 2002-12-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of non-covalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides
J.Med.Chem., 45, 2002
|
|
5A98
 
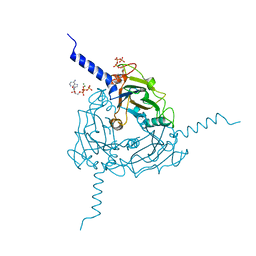 | | Crystal structure of Trichoplusia ni CPV15 polyhedra | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
1LJR
 
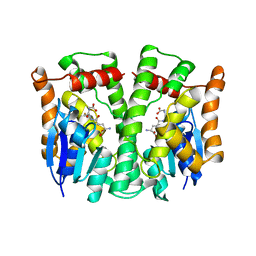 | | GLUTATHIONE TRANSFERASE (HGST T2-2) FROM HUMAN | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Rossjohn, J, Mckinstry, W.J, Oakley, A.J, Verger, D, Flanagan, J, Chelvanayagam, G, Tan, K.L, Board, P.G, Parker, M.W. | | Deposit date: | 1998-03-08 | | Release date: | 1999-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Human theta class glutathione transferase: the crystal structure reveals a sulfate-binding pocket within a buried active site.
Structure, 6, 1998
|
|
1BW5
 
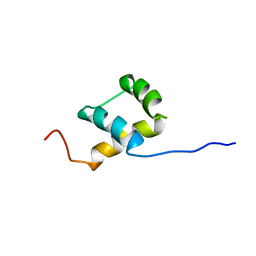 | | THE NMR SOLUTION STRUCTURE OF THE HOMEODOMAIN OF THE RAT INSULIN GENE ENHANCER PROTEIN ISL-1, 50 STRUCTURES | | Descriptor: | INSULIN GENE ENHANCER PROTEIN ISL-1 | | Authors: | Ippel, J.H, Larsson, G, Behravan, G, Zdunek, J, Lundqvist, M, Schleucher, J, Lycksell, P.-O, Wijmenga, S.S. | | Deposit date: | 1998-09-29 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the homeodomain of the rat insulin-gene enhancer protein isl-1. Comparison with other homeodomains.
J.Mol.Biol., 288, 1999
|
|
1EEM
 
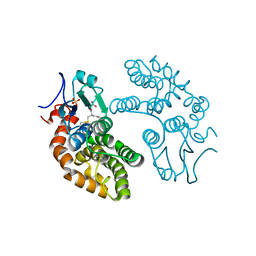 | | GLUTATHIONE TRANSFERASE FROM HOMO SAPIENS | | Descriptor: | GLUTATHIONE, GLUTATHIONE-S-TRANSFERASE, SULFATE ION | | Authors: | Board, P, Coggan, M, Chelvanayagam, G, Easteal, S, Jermiin, L.S, Schulte, G.K, Danley, D.E, Hoth, L.R, Griffor, M.C, Kamath, A.V, Rosner, M.H, Chrunyk, B.A, Perregaux, D.E, Gabel, C.A, Geoghegan, K.F, Pandit, J. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-11 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification, characterization, and crystal structure of the Omega class glutathione transferases.
J.Biol.Chem., 275, 2000
|
|
1N9A
 
 | | Farnesyltransferase complex with tetrahydropyridine inhibitors | | Descriptor: | 1-{2-[3-(4-CYANO-BENZYL)-3H-IMIDAZOL-4-YL]-ACETYL}-5-NAPHTHALEN-1-YL-1,2,3,6-TETRAHYDRO-PYRIDINE-4-CARBONITRILE, ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID, Protein farnesyltransferase alpha subunit, ... | | Authors: | Gwaltney II, S.L, O'Conner, S.J, Nelson, L.T, Sullivan, G.M, Imade, H, Wang, W, Hasvold, L, Li, Q, Cohen, J, Gu, W.Z. | | Deposit date: | 2002-11-22 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Aryl tetrahydropyridine inhibitors of farnesyltransferase: bioavailable analogues with improved cellular potency.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
4ZQX
 
 | | A revised partiality model and post-refinement algorithm for X-ray free-electron laser data | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyhedrin | | Authors: | Ginn, H.M, Brewster, A.S, Hattne, J, Evans, G, Wagner, A, Grimes, J, Sauter, N.K, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-05-11 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A revised partiality model and post-refinement algorithm for X-ray free-electron laser data.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4BBX
 
 | | Discovery of a potent, selective and orally active PDE10A inhibitor for the treatment of schizophrenia | | Descriptor: | 4-[3-[1-[(2S)-2-methoxypropyl]pyrazol-4-yl]-2-methyl-imidazo[1,2-a]pyrazin-8-yl]morpholine, CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, ... | | Authors: | Bartolome-Nebreda, J.M, Conde-Ceide, S, Delgado, F, Martin, M.L, Martinez-Viturro, C.M, Pastor, J, Tong, H.M, Iturrino, L, Macdonald, G.J, Sanderson, W, Megens, A, Langlois, X, Somers, M, Vanhoof, G. | | Deposit date: | 2012-09-28 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of a Potent, Selective and Orally Active Pde10A Inhibitor for the Potential Treatment of Schizophrenia.
J.Med.Chem., 57, 2014
|
|
5A96
 
 | | Crystal structure of Lymantria dispar CPV14 polyhedra | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-02 | | Last modified: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5A99
 
 | | Crystal structure of Operophtera brumata CPV19 polyhedra | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-02 | | Last modified: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5A8V
 
 | | Crystal structure of Orgyia pseudotsugata CPV5 polyhedra with SeMet substitution | | Descriptor: | CALCIUM ION, POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-02 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.074 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5A9P
 
 | | Crystal structure of Operophtera brumata CPV18 polyhedra | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.476 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5A8T
 
 | | Crystal structure of Antheraea mylitta CPV4 polyhedra type 2 | | Descriptor: | POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
1N94
 
 | | Aryl Tetrahydropyridine Inhbitors of Farnesyltransferase: Glycine, Phenylalanine and Histidine Derivates | | Descriptor: | 2-{(5-{[BUTYL-(2-CYCLOHEXYL-ETHYL)-AMINO]-METHYL}-2'-METHYL-BIPHENYL-2-CARBONYL)-AMINO]-4-METHYLSULFANYL-BUTYRIC ACID, ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID, Protein farnesyltransferase alpha subunit, ... | | Authors: | Gwaltney II, S.L, O'Connor, S.J, Nelson, L.T, Sullivan, G.M, Imade, H, Wang, W, Hasvold, L, Li, Q, Cohen, J, Gu, W.Z. | | Deposit date: | 2002-11-22 | | Release date: | 2003-01-07 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Aryl tetrahydropyridine inhibitors of farnesyltransferase: glycine, phenylalanine and histidine derivatives.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
5A8S
 
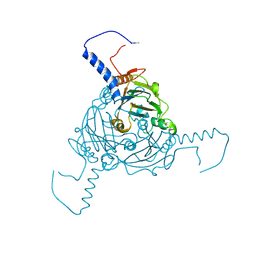 | | Crystal structure of Antheraea mylitta CPV4 polyhedra type 1 | | Descriptor: | POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.724 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
4YCR
 
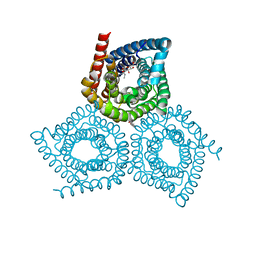 | | Structure determination of an integral membrane protein at room temperature from crystals in situ | | Descriptor: | Tellurite resistance protein TehA homolog, octyl beta-D-glucopyranoside | | Authors: | Axford, D, Hu, N.J, Foadi, J, Choudhury, H.G, Iwata, S, Beis, K, Evans, G, Alguel, Y. | | Deposit date: | 2015-02-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure determination of an integral membrane protein at room temperature from crystals in situ.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5A8U
 
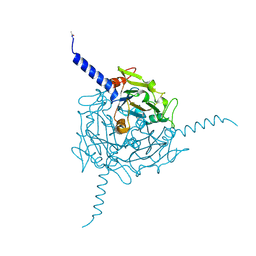 | | Crystal structure of Orgyia pseudotsugata CPV5 polyhedra | | Descriptor: | POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.609 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
1N95
 
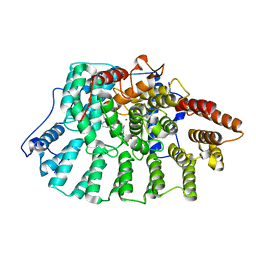 | | Aryl Tetrahydrophyridine Inhbitors of Farnesyltranferase: Glycine, Phenylalanine and Histidine Derivatives | | Descriptor: | 1-[2-(4-CYANO-BENZYLAMINO)-3-(3-METHYL-3H-IMIDAZOL-4-YL)-PROPIONYL]-5-NAPHTHALEN-1-YL-1,2,3,6-TETRAHYDRO-PYRIDINE-4-CARBONITRILE, ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID, Protein farnesyltransferase alpha subunit, ... | | Authors: | Gwaltney II, S.L, O'Conner, S.J, Nelson, L.T, Sullivan, G.M, Imade, H, Wang, W, Hasvold, L, Li, Q, Cohen, J, Gu, W.Z. | | Deposit date: | 2002-11-22 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Aryl tetrahydropyridine inhibitors of farnesyltransferase: glycine, phenylalanine and histidine derivatives.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
