4GHS
 
 | | Crystal structure of human GLTP bound with 12:0 disulfatide (orthorombic form; two subunits in asymmetric unit) | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-1-[(3,6-di-O-sulfo-beta-D-galactopyranosyl)oxy]-3-hydroxyoctadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Popov, A.N, Goni-de-Cerio, F, Cabo-Bilbao, A, Malinina, L. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GHP
 
 | | Crystal Structure of D48V||A47D mutant of Human GLTP bound with 12:0 monosulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | Deposit date: | 2012-08-08 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GJQ
 
 | | Crystal structure of human GLTP bound with 12:0 monosulfatide (orthorhombic form;two subunits in asymmetric unit) | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein, HEXANE | | Authors: | Cabo-Bilbao, A, Goni-de-Cerio, F, Samygina, V.R, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | Deposit date: | 2012-08-10 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GVT
 
 | | Crystal structure of D48V mutant of human GLTP bound with 12:0 disulfatide (hexagonal form) | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-1-[(3,6-di-O-sulfo-beta-D-galactopyranosyl)oxy]-3-hydroxyoctadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Cabo-Bilbao, A, Goni-de-Cerio, F, Popov, A.N, Malinina, L. | | Deposit date: | 2012-08-31 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GXG
 
 | | Crystal structure of human GLTP bound with 12:0 monosulfatide (orthorhombic form; four subunits in asymmetric unit) | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Cabo-Bilbao, A, Goni-de-Cerio, F, Popov, A.N, Malinina, L. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GXD
 
 | | Crystal structure of D48V mutant of human GLTP bound with 12:0 disulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-1-[(3,6-di-O-sulfo-beta-D-galactopyranosyl)oxy]-3-hydroxyoctadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4H2Z
 
 | | Crystal structure of human GLTP bound with 12:0 monosulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide, PENTADECANE, ... | | Authors: | Samygina, V.R, Ochoa-Lizarralde, B, Malinina, L. | | Deposit date: | 2012-09-13 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GH0
 
 | | Crystal structure of D48V mutant of human GLTP bound with 12:0 monosulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide, PENTADECANE | | Authors: | Samygina, V.R, Cabo-Bilbao, A, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | Deposit date: | 2012-08-07 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2MMI
 
 | | Mengovirus Leader: Structural Characterization of the Mengovirus Leader Protein Bound to Ran GTPase by Nuclear Magnetic Resonance | | Descriptor: | Leader protein, ZINC ION | | Authors: | Bacot-Davis, V.R, Cornilescu, C.C, Markley, J.L, Palmenberg, A.C. | | Deposit date: | 2014-03-15 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of Mengovirus Leader protein, its phosphorylated derivatives, and in complex with nuclear transport regulatory protein, RanGTPase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2MML
 
 | | T47 phosphorylation of the Mengovirus Leader Protein: NMR Studies of the Phosphorylation of the Mengovirus Leader Protein Reveal Stabilization of Intermolecular Domain Interactions | | Descriptor: | Leader protein, ZINC ION | | Authors: | Bacot-Davis, V.R, Porter, F.W, Palmenberg, A.C. | | Deposit date: | 2014-03-15 | | Release date: | 2014-10-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structures of Mengovirus Leader protein, its phosphorylated derivatives, and in complex with nuclear transport regulatory protein, RanGTPase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2MMK
 
 | | Y41 and T47 phosphorylation of the Mengovirus Leader Protein: NMR Studies of the Phosphorylation of the Mengovirus Leader Protein Reveal Stabilization of Intermolecular Domain Interactions | | Descriptor: | Leader protein, ZINC ION | | Authors: | Bacot-Davis, V.R, Porter, F.W, Palmenberg, A.C. | | Deposit date: | 2014-03-15 | | Release date: | 2014-10-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structures of Mengovirus Leader protein, its phosphorylated derivatives, and in complex with nuclear transport regulatory protein, RanGTPase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2N1B
 
 | |
6TPD
 
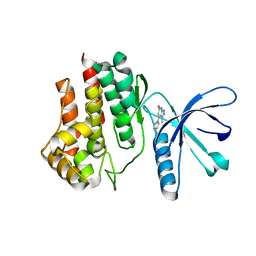 | | Fragment-based discovery of pyrazolopyridones as JAK1 inhibitors with excellent subtype selectivity | | Descriptor: | 3-methyl-4-phenyl-2,7-dihydropyrazolo[3,4-b]pyridin-6-one, Tyrosine-protein kinase JAK2 | | Authors: | Hansen, B.B, Jepsen, T.J, Larsen, M, Sindet, R, Vifian, T, Burhardt, M.N, Larsen, J, Seitzberg, J.G, Carnerup, M.A, Jerre, A, Moelck, C, Rai, S, Nasipireddy, V.R, Ritzen, A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Fragment-Based Discovery of Pyrazolopyridones as JAK1 Inhibitors with Excellent Subtype Selectivity.
J.Med.Chem., 63, 2020
|
|
6TPE
 
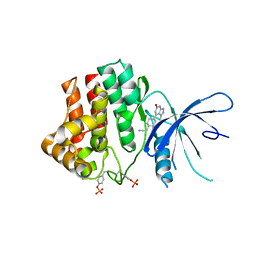 | | Fragment-based discovery of pyrazolopyridones as JAK1 inhibitors with excellent subtype selectivity | | Descriptor: | 2-[4-(3-methyl-6-oxidanylidene-1,7-dihydropyrazolo[3,4-b]pyridin-4-yl)cyclohexyl]ethanenitrile, Tyrosine-protein kinase JAK1 | | Authors: | Hansen, B.B, Jepsen, T.H, Larsen, M, Sindet, R, Vifian, T, Burhardt, M.N, Larsen, J, Seitzberg, J.G, Carnerup, M.A, Jerre, A, Molck, C, Rai, S, Nasipireddy, V.R, Jestel, A, Lammens, A, Ritzen, A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-06-10 | | Last modified: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Fragment-Based Discovery of Pyrazolopyridones as JAK1 Inhibitors with Excellent Subtype Selectivity.
J.Med.Chem., 63, 2020
|
|
6TPF
 
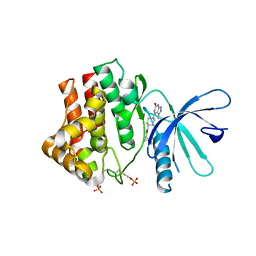 | | Fragment-based discovery of pyrazolopyridones as JAK1 inhibitors with excellent subtype selectivity | | Descriptor: | (1~{S})-2,2-bis(fluoranyl)-~{N}-[4-(3-methyl-6-oxidanylidene-2,7-dihydropyrazolo[3,4-b]pyridin-4-yl)cyclohexyl]cyclopropane-1-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Hansen, B.B, Jepsen, T.H, Larsen, M, Sindet, R, Vifian, T, Burhardt, M.N, Larsen, J, Seitzberg, J.G, Carnerup, M.A, Jerre, A, Molck, C, Rai, S, Nasipireddy, V.R, Griessner, A, Ritzen, A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-06-10 | | Last modified: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Fragment-Based Discovery of Pyrazolopyridones as JAK1 Inhibitors with Excellent Subtype Selectivity.
J.Med.Chem., 63, 2020
|
|
2MMH
 
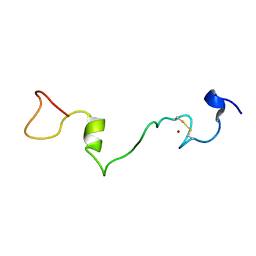 | | Unphosphorylated Mengovirus Leader Protein: NMR Studies of the Phosphorylation of the Mengovirus Leader Protein Reveal Stabilization of Intermolecular Domain Interactions | | Descriptor: | Leader protein, ZINC ION | | Authors: | Bacot-Davis, V.R, Porter, F.W, Palmenberg, A.C. | | Deposit date: | 2014-03-15 | | Release date: | 2014-10-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structures of Mengovirus Leader protein, its phosphorylated derivatives, and in complex with nuclear transport regulatory protein, RanGTPase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2MMC
 
 | | Nucleotide-free human ran gtpase | | Descriptor: | GTP-binding nuclear protein Ran | | Authors: | Bacot-Davis, V.R, Palmenberg, A.C. | | Deposit date: | 2014-03-13 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structure of Ran GTPase Determines C-terminal Tail Conformational Dynamics.
To be Published
|
|
2MMG
 
 | | Structural Characterization of the Mengovirus Leader Protein Bound to Ran GTPase by Nuclear Magnetic Resonance | | Descriptor: | GTP-binding nuclear protein Ran | | Authors: | Bacot-Davis, V.R, Palmenberg, A.C, Cornilescu, C.C, Markley, J.L. | | Deposit date: | 2014-03-15 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of Mengovirus Leader protein, its phosphorylated derivatives, and in complex with nuclear transport regulatory protein, RanGTPase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2LXY
 
 | | NMR structure of 2-MERCAPTOPHENOL-ALPHA3C | | Descriptor: | 2-MERCAPTOPHENOL, 2-mercaptophenol-alpha3C | | Authors: | Tommos, C, Valentine, K.G, Martinez-Rivera, M.C, Liang, L, Moorman, V.R. | | Deposit date: | 2012-09-06 | | Release date: | 2013-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Reversible phenol oxidation and reduction in the structurally well-defined 2-Mercaptophenol-alpha(3)C protein.
Biochemistry, 52, 2013
|
|
5KDI
 
 | | How FAPP2 Selects Simple Glycosphingolipids Using the GLTP-fold | | Descriptor: | (~{Z})-~{N}-[(~{E},2~{S},3~{R})-1-[(2~{R},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-octadec-4-en-2-yl]octadec-9-enamide, Pleckstrin homology domain-containing family A member 8 | | Authors: | Ochoa-Lizarralde, B, Popov, A.N, Samygina, V.R, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2016-06-08 | | Release date: | 2017-12-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural analyses of 4-phosphate adaptor protein 2 yield mechanistic insights into sphingolipid recognition by the glycolipid transfer protein family.
J.Biol.Chem., 293, 2018
|
|
5L5N
 
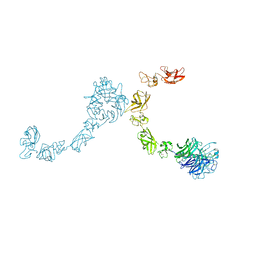 | | Plexin A4 full extracellular region, domains 1 to 7 modeled, data to 8.5 angstrom, spacegroup P4(3)22 | | Descriptor: | Plexin-A4 | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (8.502 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L5G
 
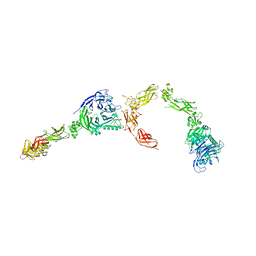 | | Plexin A2 full extracellular region, domains 1 to 8 modeled, data to 10 angstrom | | Descriptor: | Plexin-A2 | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (10 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L5M
 
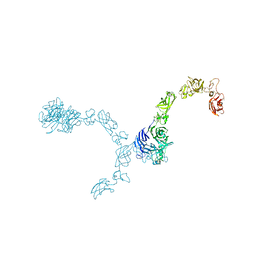 | | Plexin A4 full extracellular region, domains 1 to 7 modeled, data to 8 angstrom, spacegroup P4(3)2(1)2 | | Descriptor: | Plexin-A4 | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (8 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
6OWN
 
 | |
5L74
 
 | | Plexin A2 extracellular segment domains 4-5 (PSI2-IPT2), resolution 1.36 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Plexin-A2, ... | | Authors: | Kong, Y, Janssen, B.J.C, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-06-01 | | Release date: | 2017-03-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
