8CUR
 
 | | Crystal structure of Cdk2 in complex with Cyclin A inhibitor 6-[(E)-2-(4-chlorophenyl)ethenyl]-2-{[(2R)-3-(4-hydroxyphenyl)-1-methoxy-1-oxopropan-2-yl]carbamoyl}quinoline-4-carboxylic acid | | Descriptor: | 6-[(E)-2-(4-chlorophenyl)ethenyl]-2-{[(2R)-3-(4-hydroxyphenyl)-1-methoxy-1-oxopropan-2-yl]carbamoyl}quinoline-4-carboxylic acid, Cyclin-dependent kinase 2 | | Authors: | Tripathi, S.M, Tambo, C.S, Kiss, G, Rubin, S.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biolayer Interferometry Assay for Cyclin-Dependent Kinase-Cyclin Association Reveals Diverse Effects of Cdk2 Inhibitors on Cyclin Binding Kinetics.
Acs Chem.Biol., 18, 2023
|
|
6U1Q
 
 | | Crystal Structure of VpsO (VC0937) Kinase domain | | Descriptor: | O-PHOSPHOTYROSINE, VpsO | | Authors: | Tripathi, S.M, Schwechheimer, C, Herbert, K, Porcella, M.E, Brown, E.R, Yildiz, F.H, Rubin, S.M. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | A tyrosine phosphoregulatory system controls exopolysaccharide biosynthesis and biofilm formation in Vibrio cholerae.
Plos Pathog., 16, 2020
|
|
6U1P
 
 | | Crystal structure of VpsU (VC0916) from Vibrio cholerae | | Descriptor: | GLYCEROL, Low molecular weight phosphotyrosine protein phosphatase | | Authors: | Tripathi, S.M, Schwechheimer, C, Herbert, K, Osorio, J, Yildiz, F.H, Rubin, S.M. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | A tyrosine phosphoregulatory system controls exopolysaccharide biosynthesis and biofilm formation in Vibrio cholerae.
Plos Pathog., 16, 2020
|
|
2VOE
 
 | | Crystal structure of Rv2780 from M. tuberculosis H37Rv | | Descriptor: | ALANINE DEHYDROGENASE | | Authors: | Tripathi, S.M, Ramachandran, R. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the Mycobacterium Tuberculosis Secretory Antigen Alanine Dehydrogenase (Rv2780) in Apo and Ternary Complex Forms Captures "Open" and "Closed" Enzyme Conformations.
Proteins, 72, 2008
|
|
2VOJ
 
 | | Ternary complex of M. tuberculosis Rv2780 with NAD and pyruvate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, ALANINE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tripathi, S.M, Ramachandran, R. | | Deposit date: | 2008-02-18 | | Release date: | 2008-03-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the Mycobacterium Tuberculosis Secretory Antigen Alanine Dehydrogenase (Rv2780) in Apo and Ternary Complex Forms Captures "Open" and "Closed" Enzyme Conformations.
Proteins: Struct., Funct., Bioinf., 72, 2008
|
|
2JJF
 
 | | N328A mutant of M. tuberculosis Rv3290c | | Descriptor: | L-LYSINE EPSILON AMINOTRANSFERASE | | Authors: | tripathi, S.M, Ramachandran, R. | | Deposit date: | 2008-04-04 | | Release date: | 2009-06-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mutational Analysis of Mycobacterium Tuberculosis Lysine Epsilon-Aminotransferase and Inhibitor Co-Crystal Structures, Reveals Distinct Binding Modes.
Biochem.Biophys.Res.Commun., 463, 2015
|
|
2JJH
 
 | | E243 mutant of M. tuberculosis Rv3290C | | Descriptor: | 2-OXOGLUTARIC ACID, L-LYSINE EPSILON AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tripathi, S.M, Ramachandran, R. | | Deposit date: | 2008-04-04 | | Release date: | 2009-06-30 | | Last modified: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mutational Analysis of Mycobacterium Tuberculosis Lysine Epsilon-Aminotransferase and Inhibitor Co-Crystal Structures, Reveals Distinct Binding Modes.
Biochem.Biophys.Res.Commun., 463, 2015
|
|
2JJG
 
 | |
2JJE
 
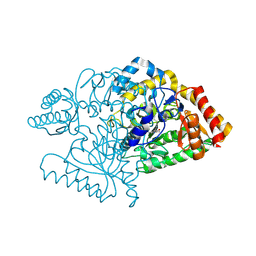 | |
2CIN
 
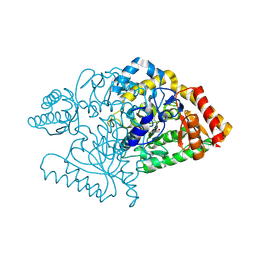 | |
2CJH
 
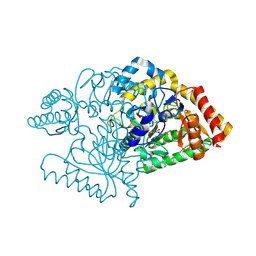 | |
2CJG
 
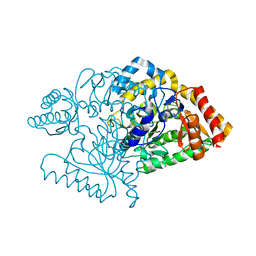 | | Lysine aminotransferase from M. tuberculosis in bound PMP form | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, L-LYSINE-EPSILON AMINOTRANSFERASE | | Authors: | Tripathi, S.M, Ramachandran, R. | | Deposit date: | 2006-04-01 | | Release date: | 2006-08-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Direct Evidence for a Glutamate Switch Necessary for Substrate Recognition: Crystal Structures of Lysine Epsilon-Aminotransferase (Rv3290C) from Mycobacterium Tuberculosis H37Rv.
J.Mol.Biol., 362, 2006
|
|
2CJD
 
 | |
6CPB
 
 | |
4IMH
 
 | |
4JX1
 
 | | Crystal structure of reduced Cytochrome P450cam-putidaredoxin complex bound to camphor and 5-exo-hydroxycamphor | | Descriptor: | 1,1'-hexane-1,6-diyldipyrrolidine-2,5-dione, 5-EXO-HYDROXYCAMPHOR, CALCIUM ION, ... | | Authors: | Tripathi, S.M, Li, H, Poulos, T.L. | | Deposit date: | 2013-03-27 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.087 Å) | | Cite: | Structural basis for effector control and redox partner recognition in cytochrome P450.
Science, 340, 2013
|
|
4JWS
 
 | | Crystal structure of Cytochrome P450cam-putidaredoxin complex | | Descriptor: | 1,1'-hexane-1,6-diyldipyrrolidine-2,5-dione, CALCIUM ION, Camphor 5-monooxygenase, ... | | Authors: | Tripathi, S.M, Li, H, Poulos, T.L. | | Deposit date: | 2013-03-27 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for effector control and redox partner recognition in cytochrome P450.
Science, 340, 2013
|
|
4JWU
 
 | | Crystal structure of Cytochrome P450cam-putidaredoxin complex | | Descriptor: | 1,1'-hexane-1,6-diyldipyrrolidine-2,5-dione, CALCIUM ION, Camphor 5-monooxygenase, ... | | Authors: | Tripathi, S.M, Li, H, Poulos, T.L. | | Deposit date: | 2013-03-27 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for effector control and redox partner recognition in cytochrome P450.
Science, 340, 2013
|
|
8GCG
 
 | | MDM2 bound to inhibitor | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, macrocyclic peptide inhibitor | | Authors: | Silvestri, A.P, Muir, E.W, Chakka, S.K, Tripathi, S.M, Rubin, S.M, Pye, C.R, Schwochert, J.A. | | Deposit date: | 2023-03-01 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | DNA-Encoded Macrocyclic Peptide Libraries Enable the Discovery of a Neutral MDM2-p53 Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
7N40
 
 | | Crystal structure of LIN9-RbAp48-LIN37, a MuvB subcomplex | | Descriptor: | Histone-binding protein RBBP4, Isoform 2 of Protein lin-9 homolog, Protein lin-37 homolog | | Authors: | Asthana, A, Ramanan, P, Tripathi, S.M, Rubin, S.M. | | Deposit date: | 2021-06-02 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The MuvB complex binds and stabilizes nucleosomes downstream of the transcription start site of cell-cycle dependent genes.
Nat Commun, 13, 2022
|
|
6P8F
 
 | | Crystal structure of CDK4 in complex with CyclinD1 and P27 | | Descriptor: | Cyclin-dependent kinase 4, Cyclin-dependent kinase inhibitor 1B, G1/S-specific cyclin-D1 | | Authors: | Guiley, K.Z, Stevenson, J.W, Lou, K, Barkovich, K.J, Bunch, K, Tripathi, S.M, Shokat, K.M, Rubin, S.M. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition.
Science, 366, 2019
|
|
6P8H
 
 | | Crystal structure of CDK4 in complex with CyclinD1 and P21 | | Descriptor: | Cyclin-dependent kinase 4, Cyclin-dependent kinase inhibitor 1, G1/S-specific cyclin-D1 | | Authors: | Guiley, K.Z, Stevenson, J.W, Lou, K, Barkovich, K.J, Bunch, K, Tripathi, S.M, Shokat, K.M, Rubin, S.M. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition.
Science, 366, 2019
|
|
6P8G
 
 | | Crystal structure of CDK4 in complex with CyclinD1 and P27 | | Descriptor: | Cyclin-dependent kinase 4, Cyclin-dependent kinase inhibitor 1B, G1/S-specific cyclin-D1 | | Authors: | Guiley, K.Z, Stevenson, J.W, Lou, K, Barkovich, K.J, Bunch, K, Tripathi, S.M, Shokat, K.M, Rubin, S.M. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition.
Science, 366, 2019
|
|
6P8E
 
 | | Crystal structure of CDK4 in complex with CyclinD1 and P27 | | Descriptor: | Cyclin-dependent kinase 4, Cyclin-dependent kinase inhibitor 1B, G1/S-specific cyclin-D1, ... | | Authors: | Guiley, K.Z, Stevenson, J.W, Lou, K, Barkovich, K.J, Bunch, K, Tripathi, S.M, Shokat, K.M, Rubin, S.M. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition.
Science, 366, 2019
|
|
8D7M
 
 | | Human Casein kinase 1 delta in complex with phosphorylated human PERIOD2 FASP peptide | | Descriptor: | Casein kinase I isoform delta, Period circadian protein homolog 2 peptide | | Authors: | Philpott, J.M, Freeberg, A.M, Tripathi, S.M, Partch, C.L. | | Deposit date: | 2022-06-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | PERIOD phosphorylation leads to feedback inhibition of CK1 activity to control circadian period.
Mol.Cell, 83, 2023
|
|
