7ZTH
 
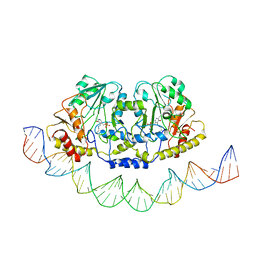 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the open conformation | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-05-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZN5
 
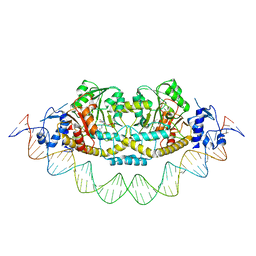 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C2 symmetry. | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-04-20 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZPA
 
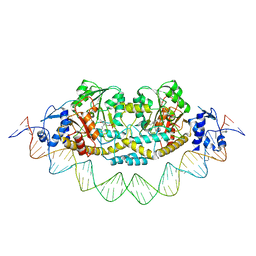 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C1 symmetry | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-04-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZLA
 
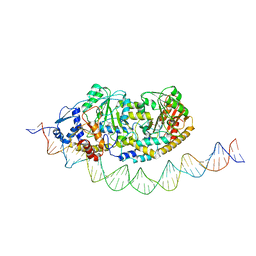 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the half-closed conformation | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Savino, C, Exertier, C, Bolognesi, M, Chaves Sanjuan, A. | | Deposit date: | 2022-04-14 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
6YMD
 
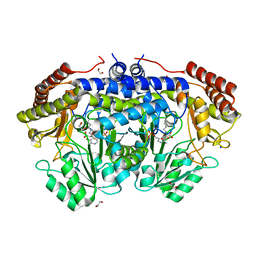 | | Crystal structure of serine hydroxymethyltransferase from Aphanothece halophytica in the covalent complex with malonate | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, MALONATE ION, ... | | Authors: | Ruszkowski, M, Sekula, B, Nogues, I, Tramonti, A, Angelaccio, S, Contestabile, R. | | Deposit date: | 2020-04-08 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural and kinetic properties of serine hydroxymethyltransferase from the halophytic cyanobacterium Aphanothece halophytica provide a rationale for salt tolerance.
Int.J.Biol.Macromol., 159, 2020
|
|
6YME
 
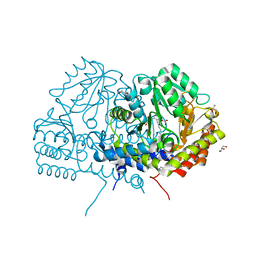 | | Crystal structure of serine hydroxymethyltransferase from Aphanothece halophytica in the PLP-internal aldimine state | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Serine hydroxymethyltransferase | | Authors: | Ruszkowski, M, Sekula, B, Nogues, I, Tramonti, A, Angelaccio, S, Contestabile, R. | | Deposit date: | 2020-04-08 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and kinetic properties of serine hydroxymethyltransferase from the halophytic cyanobacterium Aphanothece halophytica provide a rationale for salt tolerance.
Int.J.Biol.Macromol., 159, 2020
|
|
6YMF
 
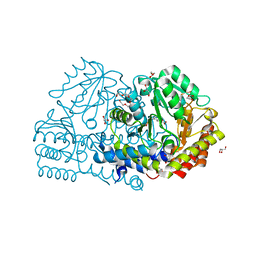 | | Crystal structure of serine hydroxymethyltransferase from Aphanothece halophytica in the PLP-Serine external aldimine state | | Descriptor: | GLYCEROL, PENTAETHYLENE GLYCOL, Serine hydroxymethyltransferase, ... | | Authors: | Ruszkowski, M, Sekula, B, Nogues, I, Tramonti, A, Angelaccio, S, Contestabile, R. | | Deposit date: | 2020-04-08 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural and kinetic properties of serine hydroxymethyltransferase from the halophytic cyanobacterium Aphanothece halophytica provide a rationale for salt tolerance.
Int.J.Biol.Macromol., 159, 2020
|
|
6YLZ
 
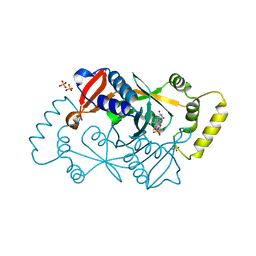 | | X-ray structure of the K72I,Y129F,R133L, H199A quadruple mutant of PNP-oxidase from E. coli | | Descriptor: | FLAVIN MONONUCLEOTIDE, PHOSPHATE ION, Pyridoxine/pyridoxamine 5'-phosphate oxidase, ... | | Authors: | Battista, T, Sularea, M, Barile, A, Fiorillo, A, Tramonti, A, Ilari, A. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.558 Å) | | Cite: | Identification and characterization of the pyridoxal 5'-phosphate allosteric site in Escherichia coli pyridoxine 5'-phosphate oxidase.
J.Biol.Chem., 296, 2021
|
|
6YMH
 
 | | X-ray structure of the K72I, Y129F, R133L, H199A quadruple mutant of PNP-oxidase from E. coli in complex with PLP | | Descriptor: | FLAVIN MONONUCLEOTIDE, PYRIDOXAL-5'-PHOSPHATE, Pyridoxine/pyridoxamine 5'-phosphate oxidase, ... | | Authors: | Battista, T, Sularea, M, Barile, A, Fiorillo, A, Tramonti, A, Ilari, A. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.417 Å) | | Cite: | Identification and characterization of the pyridoxal 5'-phosphate allosteric site in Escherichia coli pyridoxine 5'-phosphate oxidase.
J.Biol.Chem., 296, 2021
|
|
