3QUB
 
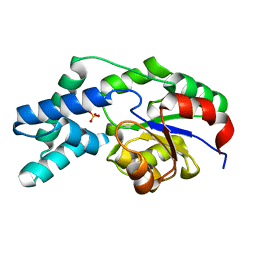 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, glu47ala mutant complexed with sulfate | | Descriptor: | INORGANIC PYROPHOSPHATASE, SULFATE ION | | Authors: | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | Deposit date: | 2011-02-23 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3QU4
 
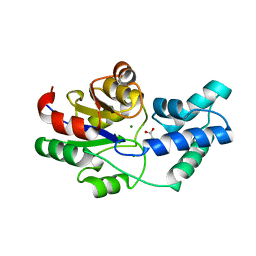 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, asp13ala mutant | | Descriptor: | ACETATE ION, CHLORIDE ION, INORGANIC PYROPHOSPHATASE, ... | | Authors: | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | Deposit date: | 2011-02-23 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3QX7
 
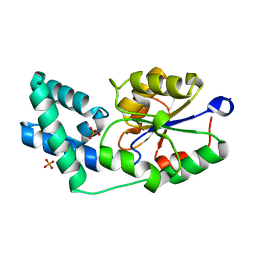 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron complexed with phosphate, a closed cap conformation | | Descriptor: | INORGANIC PYROPHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | Deposit date: | 2011-03-01 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3QUQ
 
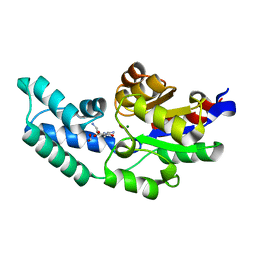 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, an open cap conformation | | Descriptor: | CHLORIDE ION, FORMIC ACID, INORGANIC PYROPHOSPHATASE, ... | | Authors: | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | Deposit date: | 2011-02-24 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3QU5
 
 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, asp11asn mutant | | Descriptor: | CHLORIDE ION, INORGANIC PYROPHOSPHATASE | | Authors: | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | Deposit date: | 2011-02-23 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3OAJ
 
 | | CRYSTAL STRUCTURE OF putative dioxygenase from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | Putative ring-cleaving dioxygenase mhqO, SULFATE ION, ZINC ION | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-08-05 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | CRYSTAL STRUCTURE OF putative dioxygenase from Bacillus subtilis subsp. subtilis str.
168
To be Published
|
|
3QU9
 
 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, asp13asn mutant complexed with magnesium and tartrate | | Descriptor: | CHLORIDE ION, GLYCEROL, INORGANIC PYROPHOSPHATASE, ... | | Authors: | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | Deposit date: | 2011-02-23 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3RCM
 
 | | crystal structure of EFI target 500140:TatD family hydrolase from Pseudomonas putida | | Descriptor: | ACETATE ION, CITRIC ACID, TatD family hydrolase, ... | | Authors: | Kim, J, Toro, R, Hillerich, B, Seidel, R.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-03-31 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | crystal structure of EFI target 500140:TatD family hydrolase from Pseudomonas putida
TO BE PUBLISHED
|
|
3RRA
 
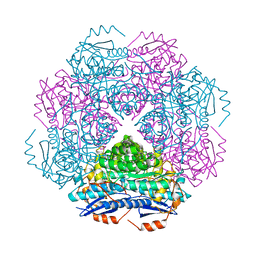 | | Crystal structure of enolase PRK14017 (target EFI-500653) from Ralstonia pickettii 12J with magnesium bound | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Putative D-galactonate dehydratase | | Authors: | Patskovsky, Y, Ramagopal, U.A, Hillerich, B, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-04-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of enolase PRK14017 from Ralstonia pickettii
To be Published
|
|
3RHG
 
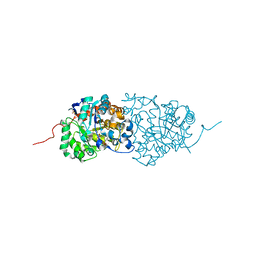 | | Crystal structure of amidohydrolase pmi1525 (target efi-500319) from proteus mirabilis hi4320 | | Descriptor: | BENZOIC ACID, CACODYLATE ION, Putative phophotriesterase, ... | | Authors: | Patskovsky, Y, Hillerich, B, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-04-11 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal Structure of Amidohydrolase Pmi1525 from Proteus Mirabilis Hi4320
To be Published
|
|
3RR1
 
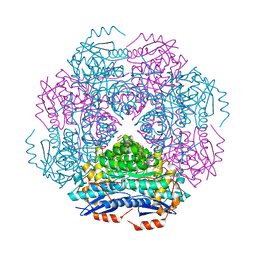 | | Crystal structure of enolase PRK14017 (target EFI-500653) from Ralstonia pickettii 12J | | Descriptor: | CHLORIDE ION, D-MALATE, Putative D-galactonate dehydratase | | Authors: | Patskovsky, Y, Hillerich, B, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-04-28 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of enolase PRK14017 from Ralstonia pickettii
To be Published
|
|
3R9K
 
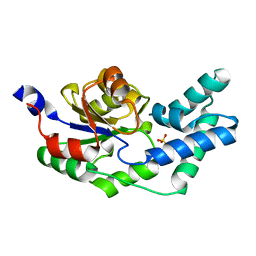 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, glu47asp mutant complexed with sulfate, a closed cap conformation | | Descriptor: | Putative beta-phosphoglucomutase, SULFATE ION | | Authors: | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | Deposit date: | 2011-03-25 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3RCY
 
 | | CRYSTAL STRUCTURE OF Mandelate racemase/muconate lactonizing enzyme-like protein from Roseovarius sp. TM1035 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme-like protein, ... | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-03-31 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | CRYSTAL STRUCTURE OF Mandelate racemase/muconate lactonizing enzyme-like protein from Roseovarius sp. TM1035
To be Published
|
|
3P9X
 
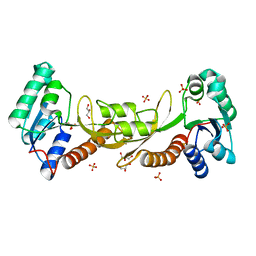 | | Crystal structure of phosphoribosylglycinamide formyltransferase from Bacillus Halodurans | | Descriptor: | GLYCEROL, SULFATE ION, phosphoribosylglycinamide formyltransferase | | Authors: | Patskovsky, Y, Toro, R, Foti, R, Seidel, R.D, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-10-18 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of phosphoribosylglycinamide formyltransferase from Bacillus Halodurans
To be Published
|
|
3SJN
 
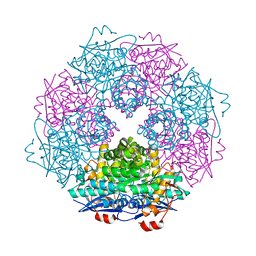 | | Crystal structure of enolase Spea_3858 (target EFI-500646) from Shewanella pealeana with magnesium bound | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein, ... | | Authors: | Patskovsky, Y, Kim, J, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Enolase Spea_3858 from Shewanella Pealeana
To be Published
|
|
2YB4
 
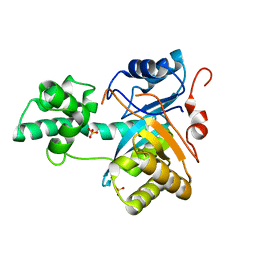 | | Structure of an amidohydrolase from Chromobacterium violaceum (EFI target EFI-500202) with bound SO4, no metal | | Descriptor: | AMIDOHYDROLASE, SULFATE ION | | Authors: | Vetting, M.W, Hillerich, B, Foti, R, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-03-01 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Prospecting for Unannotated Enzymes: Discovery of a 3',5'-Nucleotide Bisphosphate Phosphatase within the Amidohydrolase Superfamily.
Biochemistry, 53, 2014
|
|
3BSM
 
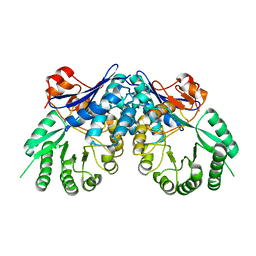 | | Crystal structure of D-mannonate dehydratase from Chromohalobacter salexigens | | Descriptor: | Mandelate racemase/muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-25 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of D-mannonate dehydratase from Chromohalobacter salexigens.
To be Published
|
|
3OP2
 
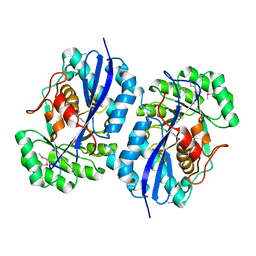 | | Crystal structure of putative mandelate racemase from Bordetella bronchiseptica RB50 complexed with 2-oxoglutarate/phosphate | | Descriptor: | 2-OXOGLUTARIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Malashkevich, V.N, Patskovsky, Y, Ramagopal, U, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative mandelate racemase from
Bordetella bronchiseptica RB50 complexed with 2-oxoglutarate/phosphate
To be Published
|
|
3OPS
 
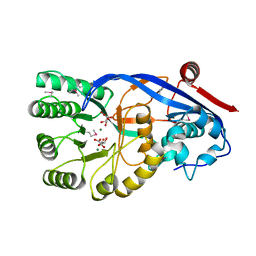 | | Crystal structure of mandelate racemase/muconate lactonizing protein FROM GEOBACILLUS SP. Y412MC10 complexed with magnesium/tartrate | | Descriptor: | D(-)-TARTARIC ACID, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Malashkevich, V.N, Patskovsky, Y, Ramagopal, U, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-01 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure ofmandelate racemase/muconate lactonizing protein FROM GEOBACILLUS SP. Y412MC10 complexed with magnesium/tartrate
To be Published
|
|
3OOQ
 
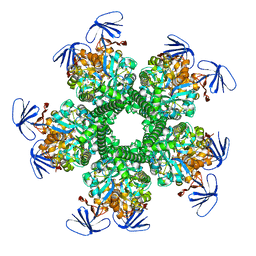 | | CRYSTAL STRUCTURE OF amidohydrolase from Thermotoga maritima MSB8 | | Descriptor: | GLYCEROL, amidohydrolase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-15 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | CRYSTAL STRUCTURE OF amidohydrolase from Thermotoga maritima MSB8
To be Published
|
|
3DDM
 
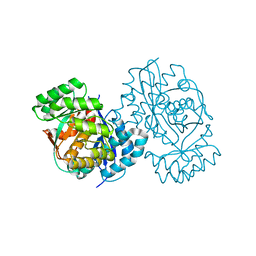 | | CRYSTAL STRUCTURE OF MANDELATE RACEMASE/MUCONATE LACTONIZING ENZYME FROM Bordetella bronchiseptica RB50 | | Descriptor: | Putative mandelate racemase/muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-05 | | Release date: | 2008-06-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CRYSTAL STRUCTURE OF MANDELATE RACEMASE/MUCONATE
LACTONIZING ENZYME FROM Bordetella bronchiseptica RB50
To be Published
|
|
2YB1
 
 | | Structure of an amidohydrolase from Chromobacterium violaceum (EFI target EFI-500202) with bound Mn, AMP and phosphate. | | Descriptor: | ADENOSINE MONOPHOSPHATE, AMIDOHYDROLASE, MANGANESE (II) ION, ... | | Authors: | Vetting, M.W, Hillerich, B, Foti, R, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-02-25 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Prospecting for Unannotated Enzymes: Discovery of a 3',5'-Nucleotide Bisphosphate Phosphatase within the Amidohydrolase Superfamily.
Biochemistry, 53, 2014
|
|
3OQB
 
 | | CRYSTAL STRUCTURE OF putative oxidoreductase from Bradyrhizobium japonicum USDA 110 | | Descriptor: | Oxidoreductase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-02 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CRYSTAL STRUCTURE OF putative oxidoreductase from Bradyrhizobium japonicum USDA 110
To be Published
|
|
3PR8
 
 | | Structure of Glutathione S-transferase(PP0183) from Pseudomonas putida in complex with GSH | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLUTATHIONE, Glutathione S-transferase family protein | | Authors: | Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-11-29 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Glutathione S-transferase(PP0183) from Pseudomonas putida in complex with GSH
To be Published
|
|
3PTW
 
 | | CRYSTAL STRUCTURE OF malonyl CoA-acyl carrier protein transacylase from Clostridium perfringens Atcc 13124 | | Descriptor: | Malonyl CoA-acyl carrier protein transacylase | | Authors: | Malashkevich, V.N, Toro, R, Ramagopal, U, Seidel, R, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-12-03 | | Release date: | 2010-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRYSTAL STRUCTURE OF malonyl CoA-acyl carrier protein transacylase from Clostridium perfringens
Atcc 13124
To be Published
|
|
