4EUN
 
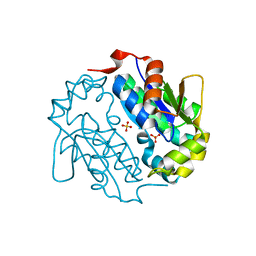 | | Crystal structure of a sugar kinase (Target EFI-502144 from Janibacter sp. HTCC2649), unliganded structure | | Descriptor: | SULFATE ION, thermoresistant glucokinase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a sugar kinase (Target EFI-502144 from Janibacter sp. HTCC2649), unliganded structure
To be Published
|
|
4E4F
 
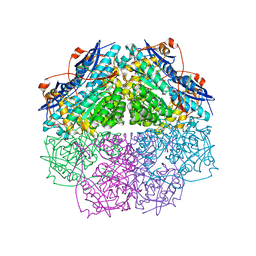 | | Crystal structure of enolase PC1_0802 (TARGET EFI-502240) from Pectobacterium carotovorum subsp. carotovorum PC1 | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-12 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of ENOLASE PC1_0802 from Pectobacterium carotovorum
To be Published
|
|
4EZE
 
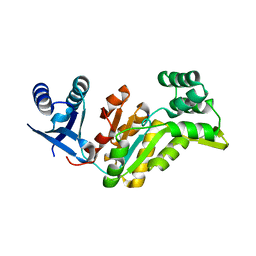 | | Crystal structure of had family hydrolase t0658 from Salmonella enterica subsp. enterica serovar Typhi (Target EFI-501419) | | Descriptor: | CHLORIDE ION, Haloacid dehalogenase-like hydrolase, SODIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-05-02 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of had hydrolase t0658 from Salmonella enterica (Target EFI-501419)
To be Published
|
|
4EZB
 
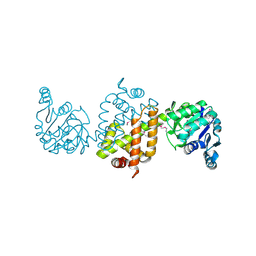 | | CRYSTAL STRUCTURE OF the Conserved hypothetical protein from Sinorhizobium meliloti 1021 | | Descriptor: | uncharacterized conserved protein | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-02 | | Release date: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRYSTAL STRUCTURE OF the Conserved hypothetical protein from Sinorhizobium meliloti 1021
To be Published
|
|
4F4R
 
 | | Crystal structure of D-mannonate dehydratase homolog from Chromohalobacter salexigens (Target EFI-502114), with bound NA, ordered loop | | Descriptor: | CHLORIDE ION, D-mannonate dehydratase, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI), Medical Structural Genomics of Pathogenic Protozoa (MSGPP) | | Deposit date: | 2012-05-11 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of D-mannonate dehydratase homolog from Chromohalobacter salexigens (Target EFI-502114), with bound NA, ordered loop
to be published
|
|
4F3X
 
 | | Crystal structure of putative aldehyde dehydrogenase from Sinorhizobium meliloti 1021 complexed with NAD | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative aldehyde dehydrogenase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-09 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of putative aldehyde dehydrogenase from Sinorhizobium meliloti 1021 complexed with NAD
To be Published
|
|
4GF0
 
 | | Crystal structure of glutahtione transferase homolog from sulfitobacter, TARGET EFI-501084, with bound glutathione | | Descriptor: | CHLORIDE ION, GLUTATHIONE, Glutathione S-transferase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-02 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of glutahtione transferase homolog from sulfitobacter, TARGET EFI-501084, with bound glutathione
To be Published
|
|
4GGH
 
 | | Crystal structure of an enolase family member from vibrio harveyi (efi-target 501692) with homology to mannonate dehydratase, with mg, hepes, and ethylene glycol bound (ordered loops, space group c2221) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-06 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an enolase family member from vibrio harveyi (efi-target 501692) with homology to mannonate dehydratase, with mg, hepes, and ethylene glycol bound (ordered loops, space group c2221)
To be Published
|
|
4G9B
 
 | | Crystal structure of beta-phosphoglucomutase homolog from escherichia coli, target efi-501172, with bound mg, open lid | | Descriptor: | Beta-phosphoglucomutase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of beta-phosphoglucomutase homolog from escherichia coli, target efi-501172, with bound mg, open lid
To be Published
|
|
4GIR
 
 | | Crystal structure of an enolase family member from vibrio harveyi (efi-target 501692) with homology to mannonate dehydratase, with mg, ethylene glycol and sulfate bound (ordered loops, space group P41212) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Enolase, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-08 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure of an enolase family member from vibrio harveyi (efi-target 501692) with homology to mannonate dehydratase, with mg, ethylene glycol and sulfate bound (ordered loops, space group P41212)
To be Published
|
|
4GME
 
 | | Crystal structure of mannonate dehydratase (target EFI-502209) from caulobacter crescentus cb15 complexed with magnesium and d-mannonate | | Descriptor: | CARBONATE ION, CHLORIDE ION, D-MANNONIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-15 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Mannonate Dehydratase from Caulobacter Crescentus Cb15
To be Published
|
|
4GLT
 
 | | Crystal structure of glutathione s-transferase MFLA_2116 (target EFI-507160) from methylobacillus flagellatus kt with gsh bound | | Descriptor: | GLUTATHIONE, Glutathione S-transferase-like protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-14 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glutathione s-transferase MFLA_2116 (target EFI-507160) from methylobacillus flagellatus kt with gsh bound
To be Published
|
|
4DNA
 
 | | CRYSTAL STRUCTURE OF putative glutathione reductase from Sinorhizobium meliloti 1021 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable glutathione reductase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-08 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CRYSTAL STRUCTURE OF putative glutathione reductase from Sinorhizobium meliloti 1021
To be Published
|
|
4DEJ
 
 | | Crystal structure of glutathione transferase-like protein IL0419 (Target EFI-501089) from Idiomarina loihiensis L2TR | | Descriptor: | Glutathione S-transferase related protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-20 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase-Like Protein Il0419 from Idiomarina Loihiensis
To be Published
|
|
4E3E
 
 | | CRYSTAL STRUCTURE OF putative MaoC domain protein dehydratase from Chloroflexus aurantiacus J-10-fl | | Descriptor: | MaoC domain protein dehydratase, SULFATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-09 | | Release date: | 2012-03-21 | | Last modified: | 2012-05-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CRYSTAL STRUCTURE OF putative MaoC domain protein dehydratase from Chloroflexus aurantiacus J-10-fl
To be Published
|
|
4DYE
 
 | | Crystal structure of an enolase (putative sugar isomerase, target efi-502095) from streptomyces coelicolor, no mg, ordered loop | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, isomerase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-28 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of an enolase (putative sugar isomerase, target efi-502095) from streptomyces coelicolor, no mg, ordered loop
to be published
|
|
4DZI
 
 | | Crystal structure of amidohydrolase map2389c (target EFI-500390) from Mycobacterium avium subsp. paratuberculosis K-10 | | Descriptor: | CADMIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-01 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of amidohydrolase map2389c (target EFI-500390) from Mycobacterium avium
To be Published
|
|
4DYK
 
 | | Crystal structure of an adenosine deaminase from pseudomonas aeruginosa pao1 (target nysgrc-200449) with bound zn | | Descriptor: | AMIDOHYDROLASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, Lafleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Zencheck, W.D, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Raushel, F.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-29 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an adenosine deaminase from pseudomonas aeruginosa pao1 (target nysgrc-200449) with bound zn
to be published
|
|
4FFU
 
 | | CRYSTAL STRUCTURE OF putative MaoC-like (monoamine oxidase-like) protein, similar to NodN from Sinorhizo Bium meliloti 1021 | | Descriptor: | CHLORIDE ION, oxidase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-06-01 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF putative MaoC-like (monoamine oxidase-like) protein, similar to NodN from Sinorhizo
Bium meliloti 1021
To be Published
|
|
4DHG
 
 | | Crystal structure of enolase TBIS_1083(TARGET EFI-502310) from Thermobispora bispora dsm 43833, an open loop conformation | | Descriptor: | GLYCEROL, IODIDE ION, Mandelate racemase/muconate lactonizing protein, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Enolase Tbis_1083 from Thermobispora Bispora Dsm 43833
To be Published
|
|
4E38
 
 | | Crystal structure of probable keto-hydroxyglutarate-aldolase from Vibrionales bacterium SWAT-3 (Target EFI-502156) | | Descriptor: | CHLORIDE ION, Keto-hydroxyglutarate-aldolase/keto-deoxy-phosphogluconate aldolase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-09 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of probable keto-hydroxyglutarate-aldolase from Vibrionales bacterium SWAT-3 (Target EFI-502156)
To be Published
|
|
4DAL
 
 | | Crystal structure of Putative aldehyde dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | GLYCEROL, Putative aldehyde dehydrogenase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-01-12 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Putative aldehyde dehydrogenase from Sinorhizobium meliloti 1021
To be Published
|
|
4FI4
 
 | | Crystal structure of mannonate dehydratase PRK15072 (TARGET EFI-502214) from Caulobacter sp. K31 | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-06-07 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Enolase Prk15072 (Target Efi-502214) from Caulobacter Sp. K31
To be Published
|
|
4E69
 
 | | Crystal structure of a sugar kinase (target EFI-502132) from Oceanicola granulosus, unliganded structure | | Descriptor: | 1,2-ETHANEDIOL, 2-dehydro-3-deoxygluconokinase, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-15 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a sugar kinase (target EFI-502132) from Oceanicola granulosus, unliganded structure
To be Published
|
|
4EEL
 
 | | Crystal structure of HAD FAMILY HYDROLASE DR_1622 from Deinococcus radiodurans R1 (TARGET EFI-501256) with bound citrate and sodium | | Descriptor: | Beta-phosphoglucomutase-related protein, CITRIC ACID, SODIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-28 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of HAD HYDROLASE DR_1622 Deinococcus radiodurans R1 (TARGET EFI-501256)
To be Published
|
|
