3IVR
 
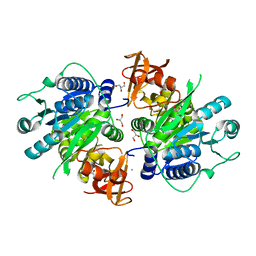 | | CRYSTAL STRUCTURE OF PUTATIVE long-chain-fatty-acid CoA ligase FROM Rhodopseudomonas palustris CGA009 | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative long-chain-fatty-acid CoA ligase | | Authors: | Patskovsky, Y, Toro, R, Foti, R, Dickey, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-01 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF PUTATIVE long-chain-fatty-acid CoA SYNTHASE FROM Rhodopseudomonas palustris CGA009
To be Published
|
|
3HCW
 
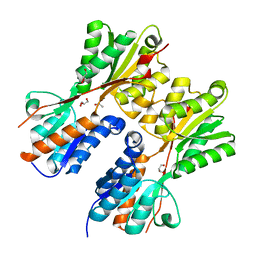 | | CRYSTAL STRUCTURE OF PROBABLE maltose operon transcriptional repressor malR FROM STAPHYLOCOCCUS AREUS | | Descriptor: | GLYCEROL, Maltose operon transcriptional repressor | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Maltose Operon Transcriptional Repressor from Staphylococcus Aureus
To be Published
|
|
3K2W
 
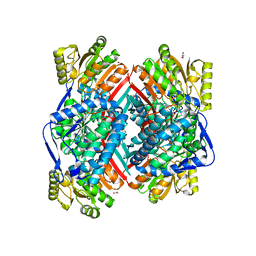 | | CRYSTAL STRUCTURE OF betaine-aldehyde dehydrogenase FROM Pseudoalteromonas atlantica T6c | | Descriptor: | ACETATE ION, Betaine-aldehyde dehydrogenase, CHLORIDE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CRYSTAL STRUCTURE OF betaine-aldehyde dehydrogenase FROM Pseudoalteromonas atlantica
To be Published
|
|
3K4U
 
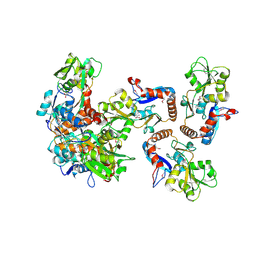 | | CRYSTAL STRUCTURE OF putative binding component of ABC transporter from Wolinella succinogenes DSM 1740 complexed with lysine | | Descriptor: | BINDING COMPONENT OF ABC TRANSPORTER, LYSINE | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-06 | | Release date: | 2009-11-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | CRYSTAL STRUCTURE OF putative binding component of ABC transporter from Wolinella succinogenes
DSM 1740 complexed with lysine
To be Published
|
|
3G7U
 
 | | Crystal structure of putative DNA modification methyltransferase encoded within prophage Cp-933R (E.coli) | | Descriptor: | CHLORIDE ION, Cytosine-specific methyltransferase, GLYCEROL | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Gilmore, M, Iizuka, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-10 | | Release date: | 2009-02-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of DNA Modification Methyltransferase Encoded within Prophage Cp-933R (E.coli)
To be Published
|
|
3GG9
 
 | | CRYSTAL STRUCTURE OF putative D-3-phosphoglycerate dehydrogenase oxidoreductase from Ralstonia solanacearum | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Putative D-3-Phosphoglycerate Dehydrogenase from Ralstonia Solanacearum
To be Published
|
|
5BR1
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS S4 (Avi_5305, TARGET EFI-511224) WITH BOUND ALPHA-D-GALACTOSAMINE | | Descriptor: | 2-amino-2-deoxy-alpha-D-galactopyranose, ABC transporter, binding protein | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-05-29 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of an ABC transporter solute-binding protein specific for the amino sugars glucosamine and galactosamine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
3GRZ
 
 | | CRYSTAL STRUCTURE OF ribosomal protein L11 methylase FROM Lactobacillus delbrueckii subsp. bulgaricus | | Descriptor: | GLYCEROL, Ribosomal protein L11 methyltransferase | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN 11 METHYLASE FROM Lactobacillus delbrueckii subsp. bulgaricus
To be Published
|
|
3H7L
 
 | | CRYSTAL STRUCTURE OF ENDOGLUCANASE-RELATED PROTEIN FROM Vibrio parahaemolyticus | | Descriptor: | ENDOGLUCANASE, GLYCEROL | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Rutter, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-27 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF ENDOGLUCANASE-RELATED PROTEIN FROM Vibrio parahaemolyticus
To be Published
|
|
3HN2
 
 | | Crystal structure of 2-dehydropantoate 2-reductase FROM Geobacter metallireducens GS-15 | | Descriptor: | 2-dehydropantoate 2-reductase | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Rutter, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-29 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 2-dehydropantoate 2-reductase FROM Geobacter metallireducens
To be Published
|
|
3HV2
 
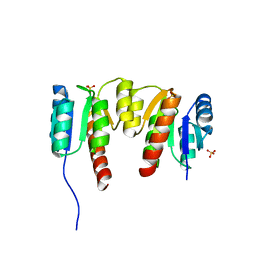 | | Crystal structure of signal receiver domain OF HD domain-containing protein FROM Pseudomonas fluorescens Pf-5 | | Descriptor: | Response regulator/HD domain protein, SULFATE ION | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-15 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of signal receiver domain oF HD domain-containing protein 3 FROM Pseudomonas fluorescens
To be Published
|
|
3JVD
 
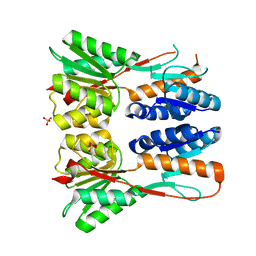 | | Crystal structure of putative transcription regulation repressor (LACI family) FROM Corynebacterium glutamicum | | Descriptor: | GLYCEROL, SULFATE ION, Transcriptional regulators | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Foti, R, Freeman, J, Do, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-16 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of LACI family protein from Corynebacterium glutamicum
To be Published
|
|
3K4H
 
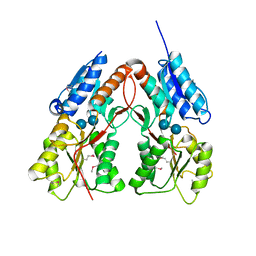 | | CRYSTAL STRUCTURE OF putative transcriptional regulator LacI from Bacillus cereus subsp. cytotoxis NVH 391-98 | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, putative transcriptional regulator | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-05 | | Release date: | 2009-11-10 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CRYSTAL STRUCTURE OF putative transcriptional regulator LacI from Bacillus cereus subsp. cytotoxis NVH 391-98
To be Published
|
|
3K4W
 
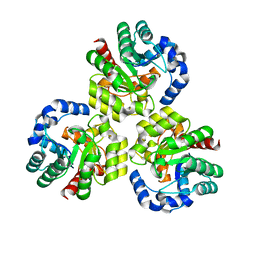 | | CRYSTAL STRUCTURE OF Uncharacterized Tim-Barrel Protein Bb4693 From Bordetella Bronchiseptica | | Descriptor: | uncharacterized protein Bb4693 | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-06 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | CRYSTAL STRUCTURE OF Uncharacterized Tim-Barrel Protein
Bb4693 From Bordetella Bronchiseptica
To be Published
|
|
3K9D
 
 | | CRYSTAL STRUCTURE OF PROBABLE ALDEHYDE DEHYDROGENASE FROM Listeria monocytogenes EGD-e | | Descriptor: | ALDEHYDE DEHYDROGENASE, CHLORIDE ION, GLYCEROL | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-15 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Aldehyde Dehydrogenase from Listeria Monocytogenes
To be Published
|
|
3GH1
 
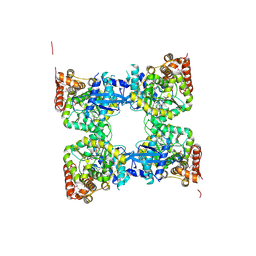 | | Crystal structure of predicted nucleotide-binding protein from Vibrio cholerae | | Descriptor: | PHOSPHATE ION, Predicted nucleotide-binding protein | | Authors: | Malashkevich, V.N, Toro, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-02 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of predicted nucleotide-binding protein from Vibrio cholerae.
To be Published
|
|
3GY1
 
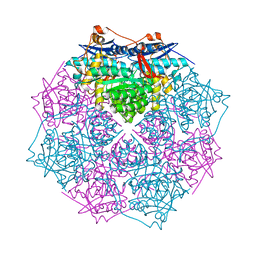 | | CRYSTAL STRUCTURE OF putative mandelate racemase/muconate lactonizing protein from Clostridium beijerinckii NCIMB 8052 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-03 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF putative mandelate racemase/muconate lactonizing protein from Clostridium
beijerinckii NCIMB 8052
To be Published
|
|
3GNL
 
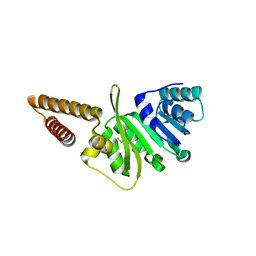 | | Structure of uncharacterized protein (LMOf2365_1472) from Listeria monocytogenes serotype 4b | | Descriptor: | uncharacterized protein, DUF633, LMOf2365_1472 | | Authors: | Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-17 | | Release date: | 2009-04-21 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of uncharacterized protein (LMOf2365_1472) from
Listeria monocytogenes serotype 4b
To be Published
|
|
3GP4
 
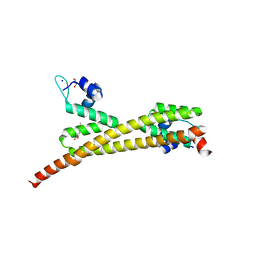 | | Crystal structure of putative MerR family transcriptional regulator from listeria monocytogenes | | Descriptor: | D-METHIONINE, GLYCEROL, SODIUM ION, ... | | Authors: | Ramagopal, U.A, Toro, R, Morano, C, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-20 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of putative MerR family transcriptional regulator from Listeria monocytogenes
To be published
|
|
3GY0
 
 | | Crystal structure of putatitve short chain dehydrogenase FROM SHIGELLA FLEXNERI 2A STR. 301 complexed with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, dehydrogenase | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-03 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of putatitve short chain dehydrogenase
FROM SHIGELLA FLEXNERI 2A STR. 301 complexed with NADP
To be Published
|
|
3HG7
 
 | | CRYSTAL STRUCTURE OF D-isomer specific 2-hydroxyacid dehydrogenase family protein from Aeromonas salmonicida subsp. salmonicida A449 | | Descriptor: | D-isomer specific 2-hydroxyacid dehydrogenase family protein, GLYCEROL | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-13 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF D-isomer specific 2-hydroxyacid dehydrogenase family protein from Aeromonas salmonicida subsp. salmonicida A449
To be Published
|
|
3HIN
 
 | | CRYSTAL STRUCTURE OF putative enoyl-CoA hydratase from Rhodopseudomonas palustris CGA009 | | Descriptor: | Putative 3-hydroxybutyryl-CoA dehydratase | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-20 | | Release date: | 2009-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF putative enoyl-CoA hydratase from Rhodopseudomonas palustris CGA009
To be Published
|
|
3G2E
 
 | | Structure of putative OORC subunit of 2-oxoglutarate:acceptor oxidoreductase from Campylobacter jejuni | | Descriptor: | GLYCEROL, OORC subunit of 2-oxoglutarate:acceptor oxidoreductase | | Authors: | Ramagopal, U.A, Toro, R, Miller, S, Gilmore, M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-31 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of putative OORC subunit of 2-oxoglutarate:acceptor oxidoreductase from
Campylobacter jejuni
To be Published
|
|
3GDO
 
 | |
3GFO
 
 | | Structure of cbiO1 from clostridium perfringens: Part of the ABC transporter complex cbiONQ. | | Descriptor: | Cobalt import ATP-binding protein cbiO 1, SULFATE ION | | Authors: | Ramagopal, U.A, Morano, C, Toro, R, Dickey, M, Do, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of cbiO1 from clostridium perfringens: Part of the ABC transporter complex cbiONQ
To be published
|
|
