3JD6
 
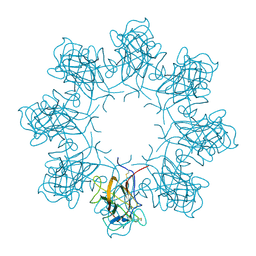 | | Double octamer structure of retinoschisin, a cell-cell adhesion protein of the retina | | Descriptor: | Retinoschisin | | Authors: | Tolun, G, Vijayasarathy, C, Huang, R, Zeng, Y, Li, Y, Steven, A.C, Sieving, P.A, Heymann, J.B. | | Deposit date: | 2016-04-12 | | Release date: | 2016-05-11 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Paired octamer rings of retinoschisin suggest a junctional model for cell-cell adhesion in the retina.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7UJL
 
 | |
6WVJ
 
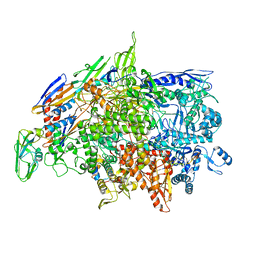 | | Cryo-EM structure of Bacillus subtilis RNA Polymerase elongation complex | | Descriptor: | DNA (5'-D(*TP*GP*TP*CP*GP*GP*GP*CP*GP*TP*CP*CP*GP*CP*GP*CP*GP*CP*C)-3'), DNA (5'-D(P*AP*CP*GP*CP*CP*CP*GP*AP*CP*A)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Newing, T, Tolun, G, Oakley, A.J. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Molecular basis for RNA polymerase-dependent transcription complex recycling by the helicase-like motor protein HelD.
Nat Commun, 11, 2020
|
|
6WVK
 
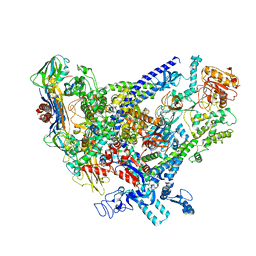 | | Cryo-EM structure of Bacillus subtilis RNA Polymerase in complex with HelD | | Descriptor: | DNA helicase IV, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Newing, T, Tolun, G, Oakley, A.J. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Molecular basis for RNA polymerase-dependent transcription complex recycling by the helicase-like motor protein HelD.
Nat Commun, 11, 2020
|
|
4D1Q
 
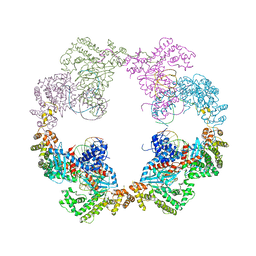 | | Hermes transposase bound to its terminal inverted repeat | | Descriptor: | SODIUM ION, TERMINAL INVERTED REPEAT, TRANSPOSASE | | Authors: | Hickman, A.B, Ewis, H, Li, X, Knapp, J, Laver, T, Doss, A.L, Tolun, G, Steven, A, Grishaev, A, Bax, A, Atkinson, P, Craig, N.L, Dyda, F. | | Deposit date: | 2014-05-04 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Hat Transposon End Recognition by Hermes, an Octameric DNA Transposase from Musca Domestica.
Cell(Cambridge,Mass.), 158, 2014
|
|
