7OD3
 
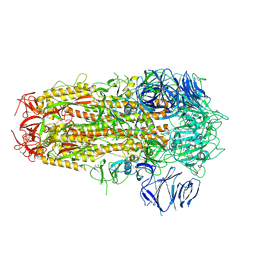 | | SARS CoV-2 Spike protein, Bristol UK Deletion variant, Closed conformation, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2021-04-28 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights in cell-type specific evolution of intra-host diversity by SARS-CoV-2.
Nat Commun, 13, 2022
|
|
7ODL
 
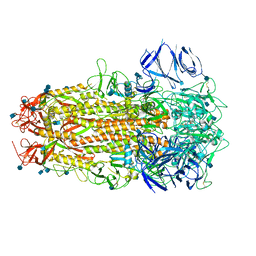 | | SARS CoV-2 Spike protein, Bristol UK Deletion variant, Closed conformation, C1 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2021-04-29 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insights in cell-type specific evolution of intra-host diversity by SARS-CoV-2.
Nat Commun, 13, 2022
|
|
7ZH2
 
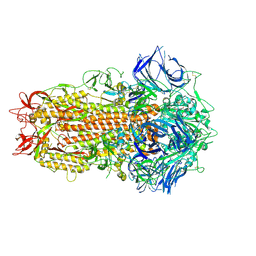 | | SARS CoV Spike protein, Closed C1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Buzas, D, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2022-04-05 | | Release date: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | The free fatty acid-binding pocket is a conserved hallmark in pathogenic beta-coronavirus spike proteins from SARS-CoV to Omicron.
Sci Adv, 8, 2022
|
|
7ZH5
 
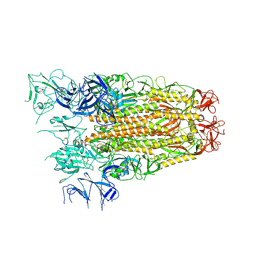 | | SARS CoV Spike protein, Open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Buzas, D, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2022-04-05 | | Release date: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The free fatty acid-binding pocket is a conserved hallmark in pathogenic beta-coronavirus spike proteins from SARS-CoV to Omicron.
Sci Adv, 8, 2022
|
|
7ZH1
 
 | | SARS CoV Spike protein, Closed C3 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Buzas, D, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2022-04-05 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | The free fatty acid-binding pocket is a conserved hallmark in pathogenic beta-coronavirus spike proteins from SARS-CoV to Omicron.
Sci Adv, 8, 2022
|
|
6ZB5
 
 | | SARS CoV-2 Spike protein, Closed conformation, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Burucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2020-06-07 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein.
Science, 370, 2020
|
|
6ZB4
 
 | | SARS CoV-2 Spike protein, Closed conformation, C1 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Burucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2020-06-06 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein.
Science, 370, 2020
|
|
6ZE0
 
 | |
6ZDZ
 
 | |
5EFZ
 
 | | Monoclinic structure of the acetyl esterase MekB | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Toelzer, C, Pal, S, Watzlawick, H, Altenbuchner, J, Niefind, K. | | Deposit date: | 2015-10-26 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A novel esterase subfamily with alpha / beta-hydrolase fold suggested by structures of two bacterial enzymes homologous to l-homoserine O-acetyl transferases.
Febs Lett., 590, 2016
|
|
6Q6Z
 
 | |
8COI
 
 | | Human adenovirus-derived synthetic ADDobody binder | | Descriptor: | ADDobody | | Authors: | Buzas, D, Toelzer, C, Gupta, K, Berger-Schaffitzel, C, Berger, I. | | Deposit date: | 2023-02-28 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Engineering the ADDobody protein scaffold for generation of high-avidity ADDomer super-binders.
Structure, 32, 2024
|
|
8QB3
 
 | | ADDobody zinc containing condition | | Descriptor: | ADDobody, ZINC ION | | Authors: | Buzas, D, Toelzer, C, Berger, I, Schaffitzel, C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering the ADDobody protein scaffold for generation of high-avidity ADDomer super-binders.
Structure, 32, 2024
|
|
8QBX
 
 | | Chimeric Adenovirus-derived dodecamer | | Descriptor: | Penton protein | | Authors: | Buzas, D, Borucu, U, Bufton, J, Kapadalakere, S.Y, Toelzer, C. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Engineering the ADDobody protein scaffold for generation of high-avidity ADDomer super-binders.
Structure, 32, 2024
|
|
6I8G
 
 | |
7A1B
 
 | | Crystal structure of human protein kinase CK2alpha' (CSNK2A2 gene product) in complex with the ATP-competitive inhibitor 5,6-dibromo-1H-triazolo[4,5-b]pyridine | | Descriptor: | 1,2-ETHANEDIOL, 5,6-dibromo-1H-triazolo[4,5-b]pyridine, CHLORIDE ION, ... | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-12 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.287 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
7A2H
 
 | | Crystal structure of human protein kinase CK2alpha' (CSNK2A2 gene product) in complex with the ATP-competitive inhibitor 5,6,7-tribromo-1H-imidazo[4,5-b]pyridine | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7-tris(bromanyl)-1~{H}-imidazo[4,5-b]pyridine, Casein kinase II subunit alpha' | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-18 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
7A4C
 
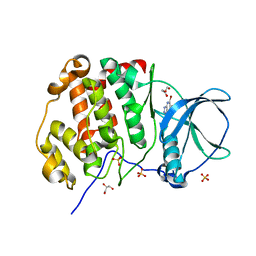 | | Crystal structure of human protein kinase CK2alpha (CSNK2A1 gene product) in complex with the ATP-competitive inhibitor 5,6,7-tribromo-1H-triazolo[4,5-b]pyridine | | Descriptor: | 5,6,7-tris(bromanyl)-1~{H}-[1,2,3]triazolo[4,5-b]pyridine, Casein kinase II subunit alpha, GLYCEROL, ... | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-19 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
7A22
 
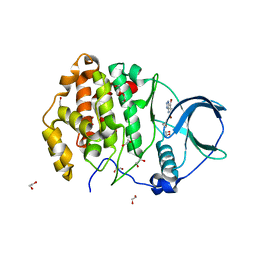 | | Crystal structure of human protein kinase CK2alpha' (CSNK2A2 gene product) in complex with the ATP-competitive inhibitor 5,6,7-tribromo-1H-triazolo[4,5-b]pyridine | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7-tris(bromanyl)-1~{H}-[1,2,3]triazolo[4,5-b]pyridine, Casein kinase II subunit alpha' | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
7A4B
 
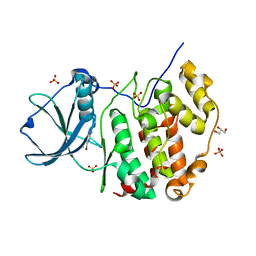 | | Crystal structure of human protein kinase CK2alpha (CSNK2A1 gene product) in complex with the ATP-competitive inhibitor 5,6-dibromo-1H-triazolo[4,5-b]pyridine | | Descriptor: | 5,6-dibromo-1H-triazolo[4,5-b]pyridine, Casein kinase II subunit alpha, GLYCEROL, ... | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-19 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
7A49
 
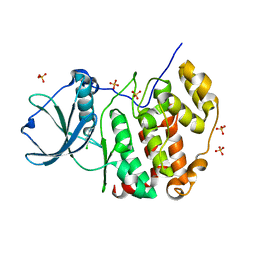 | | Crystal structure of human protein kinase CK2alpha (CSNK2A1 gene product) in complex with the ATP-competitive inhibitor 6-bromo-5-chloro-1H-triazolo[4,5-b]pyridine | | Descriptor: | 6-bromanyl-5-chloranyl-1~{H}-[1,2,3]triazolo[4,5-b]pyridine, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-19 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
6I8H
 
 | |
7A1Z
 
 | | Crystal structure of human protein kinase CK2alpha' (CSNK2A2 gene product) in complex with the ATP-competitive inhibitor 6-bromo-5-chloro-1H-triazolo[4,5-b]pyridine | | Descriptor: | 1,2-ETHANEDIOL, 6-bromanyl-5-chloranyl-1~{H}-[1,2,3]triazolo[4,5-b]pyridine, CHLORIDE ION, ... | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.024 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
5D7B
 
 | | Trigonal Crystal Structure of an acetylester hydrolase from Corynebacterium glutamicum | | Descriptor: | GLYCEROL, Homoserine O-acetyltransferase | | Authors: | Niefind, K, Toelzer, C, Pal, S, Altenbuchner, J, Watzlawick, H. | | Deposit date: | 2015-08-13 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A novel esterase subfamily with alpha / beta-hydrolase fold suggested by structures of two bacterial enzymes homologous to l-homoserine O-acetyl transferases.
Febs Lett., 590, 2016
|
|
5D6O
 
 | | Orthorhombic Crystal Structure of an acetylester hydrolase from Corynebacterium glutamicum | | Descriptor: | CHLORIDE ION, GLYCEROL, Homoserine O-acetyltransferase, ... | | Authors: | Niefind, K, Toelzer, C, Altenbuchner, J, Watzlawick, H. | | Deposit date: | 2015-08-12 | | Release date: | 2015-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel esterase subfamily with alpha / beta-hydrolase fold suggested by structures of two bacterial enzymes homologous to l-homoserine O-acetyl transferases.
Febs Lett., 590, 2016
|
|
