1DM2
 
 | | HUMAN CYCLIN-DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR HYMENIALDISINE | | Descriptor: | 1,2-ETHANEDIOL, 4-(5-AMINO-4-OXO-4H-PYRAZOL-3-YL)-2-BROMO-4,5,6,7-TETRAHYDRO-3AH-PYRROLO[2,3-C]AZEPIN-8-ONE, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Thunnissen, A.M, Kim, S.-H. | | Deposit date: | 1999-12-13 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of cyclin-dependent kinases, GSK-3beta and CK1 by hymenialdisine, a marine sponge constituent.
Chem.Biol., 7, 2000
|
|
4X1C
 
 | |
4X19
 
 | |
7P76
 
 | | Re-engineered 2-deoxy-D-ribose-5-phosphate aldolase catalysing asymmetric Michael addition reactions, Schiff base complex with cinnamaldehyde | | Descriptor: | (2E)-3-phenylprop-2-enal, Deoxyribose-phosphate aldolase, GLYCEROL | | Authors: | Thunnissen, A.M.W.H, Rozeboom, H.J, Kunzendorf, A, Poelarends, G.J. | | Deposit date: | 2021-07-19 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unlocking Asymmetric Michael Additions in an Archetypical Class I Aldolase by Directed Evolution.
Acs Catalysis, 11, 2021
|
|
7P75
 
 | |
5CLO
 
 | |
5CLN
 
 | |
1SLY
 
 | | COMPLEX OF THE 70-KDA SOLUBLE LYTIC TRANSGLYCOSYLASE WITH BULGECIN A | | Descriptor: | 4-O-(4-O-SULFONYL-N-ACETYLGLUCOSAMININYL)-5-METHYLHYDROXY-L-PROLINE-TAURINE, 70-KDA SOLUBLE LYTIC TRANSGLYCOSYLASE | | Authors: | Thunnissen, A.M.W.H, Kalk, K.H, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1995-08-02 | | Release date: | 1996-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the 70-kDa soluble lytic transglycosylase complexed with bulgecin A. Implications for the enzymatic mechanism.
Biochemistry, 34, 1995
|
|
9H9Y
 
 | | Crystal structure of metal-free LmrR_V15Bpy variant BVS in a closed state | | Descriptor: | Transcriptional regulator, PadR-like family | | Authors: | Thunnissen, A.M.W.H, Jiang, R, Casilli, F, Aalbers, F, Roelfes, G. | | Deposit date: | 2024-11-01 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An artificial copper-Michaelase featuring a genetically encoded bipyridine ligand for asymmetric additions to nitroalkenes.
Angew.Chem.Int.Ed.Engl., 2025
|
|
9HA0
 
 | | Crystal structure of Cu(II)-bound LmrR_V15Bpy variant BVS | | Descriptor: | COPPER (II) ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Thunnissen, A.M.W.H, Jiang, R, Casilli, F, Aalbers, F, Roelfes, G. | | Deposit date: | 2024-11-01 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An artificial copper-Michaelase featuring a genetically encoded bipyridine ligand for asymmetric additions to nitroalkenes.
Angew.Chem.Int.Ed.Engl., 2025
|
|
9GKT
 
 | | Crystal structure of artificial enzyme LmrR_pAF variant RGN | | Descriptor: | Transcriptional regulator, PadR-like family | | Authors: | Thunnissen, A.M.W.H, Leveson-Gower, R.B, Rozeboom, H.J, Roelfes, G. | | Deposit date: | 2024-08-26 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Evolutionary Specialization of a Promiscuous Designer Enzyme.
Acs Catalysis, 15, 2025
|
|
9FD8
 
 | | Re-engineered peroxygenase variant of 2-deoxy-D-ribose-5-phosphate aldolase, Schiff-base complex with 4-chloro-cinnamaldehyde | | Descriptor: | (2E)-3-(4-chlorophenyl)prop-2-enal, Deoxyribose-phosphate aldolase, GLYCEROL | | Authors: | Thunnissen, A.M.W.H, Zhou, H, Frietema, H.O.T, Poelarends, G.J. | | Deposit date: | 2024-05-16 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Engineering 2-Deoxy-D-ribose-5-phosphate Aldolase for anti- and syn-Selective Epoxidations of alpha , beta-Unsaturated Aldehydes.
Angew.Chem.Int.Ed.Engl., 2025
|
|
9H9W
 
 | | Crystal structure of metal-free LmrR_V15Bpy in an open state | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Transcriptional regulator, PadR-like family | | Authors: | Thunnissen, A.M.W.H, Jiang, R, Casilli, F, Aalbers, F, Roelfes, G. | | Deposit date: | 2024-11-01 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | An artificial copper-Michaelase featuring a genetically encoded bipyridine ligand for asymmetric additions to nitroalkenes.
Angew.Chem.Int.Ed.Engl., 2025
|
|
9FD7
 
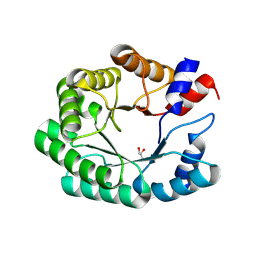 | | Re-engineered peroxygenase variant of 2-deoxy-D-ribose-5-phosphate aldolase in substrate-free state | | Descriptor: | Deoxyribose-phosphate aldolase, GLYCEROL | | Authors: | Thunnissen, A.M.W.H, Zhou, H, Frietema, H.O.T, Poelarends, G.J. | | Deposit date: | 2024-05-16 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineering 2-Deoxy-D-ribose-5-phosphate Aldolase for anti- and syn-Selective Epoxidations of alpha , beta-Unsaturated Aldehydes.
Angew.Chem.Int.Ed.Engl., 2025
|
|
9GKR
 
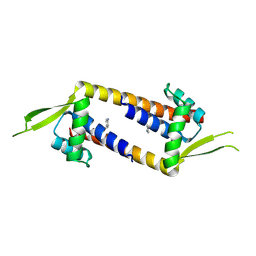 | | Crystal structure of artificial enzyme LmrR_pAF variant RMH in crystal form 1 | | Descriptor: | Transcriptional regulator, PadR-like family | | Authors: | Thunnissen, A.M.W.H, Leveson-Gower, R.B, Rozeboom, H.J, Roelfes, G. | | Deposit date: | 2024-08-26 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Evolutionary Specialization of a Promiscuous Designer Enzyme.
Acs Catalysis, 15, 2025
|
|
9GKS
 
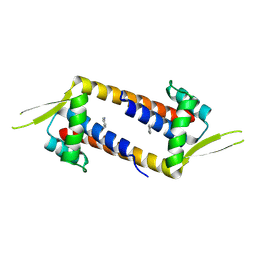 | | Crystal structure of artificial enzyme LmrR_pAF variant RMH in crystal form 2 | | Descriptor: | Transcriptional regulator, PadR-like family | | Authors: | Thunnissen, A.M.W.H, Leveson-Gower, R.B, Rozeboom, H.J, Roelfes, G. | | Deposit date: | 2024-08-26 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Evolutionary Specialization of a Promiscuous Designer Enzyme.
Acs Catalysis, 15, 2025
|
|
9FD9
 
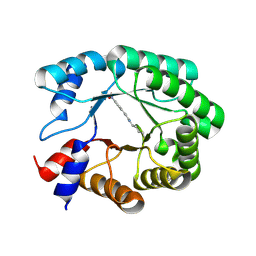 | | Re-engineered peroxygenase variant of 2-deoxy-D-ribose-5-phosphate aldolase, Schiff-base complex with 4-nitro-cinnamaldehyde | | Descriptor: | 4-nitro-cinnamaldehyde, Deoxyribose-phosphate aldolase, GLYCEROL | | Authors: | Thunnissen, A.M.W.H, Zhou, H, Frietema, H.O.T, Poelarends, G.J. | | Deposit date: | 2024-05-16 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Engineering 2-Deoxy-D-ribose-5-phosphate Aldolase for anti- and syn-Selective Epoxidations of alpha , beta-Unsaturated Aldehydes.
Angew.Chem.Int.Ed.Engl., 2025
|
|
9G52
 
 | | Crystal structure of LmrR with V15 replaced by unnatural amino acid 4-mercaptophenylalanine, Au(I) bound | | Descriptor: | GOLD ION, Transcriptional regulator, PadR-like family | | Authors: | Thunnissen, A.M.W.H, Aalbers, F.S, Veen, M.J, Rozeboom, H.J, Roelfes, G. | | Deposit date: | 2024-07-16 | | Release date: | 2024-12-25 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Artificial Gold Enzymes Using a Genetically Encoded Thiophenol-Based Noble-Metal-Binding Ligand.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9G51
 
 | | Crystal structure of LmrR with V15 replaced by unnatural amino acid 4-mercaptophenylalanine, apo | | Descriptor: | Transcriptional regulator, PadR-like family | | Authors: | Thunnissen, A.M.W.H, Aalbers, F.S, Veen, M.J, Rozeboom, H.J, Roelfes, G. | | Deposit date: | 2024-07-16 | | Release date: | 2024-12-25 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Artificial Gold Enzymes Using a Genetically Encoded Thiophenol-Based Noble-Metal-Binding Ligand.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9H9Z
 
 | | Crystal structure of Cu(II)-bound LmrR_V15Bpy | | Descriptor: | COPPER (II) ION, MALONATE ION, Transcriptional regulator, ... | | Authors: | Thunnissen, A.M.W.H, Jiang, R, Casilli, F, Aalbers, F, Roelfes, G. | | Deposit date: | 2024-11-01 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An artificial copper-Michaelase featuring a genetically encoded bipyridine ligand for asymmetric additions to nitroalkenes.
Angew.Chem.Int.Ed.Engl., 2025
|
|
9H9X
 
 | | Crystal structure of metal-free LmrR_V15Bpy in a closed state | | Descriptor: | Transcriptional regulator, PadR-like family | | Authors: | Thunnissen, A.M.W.H, Jiang, R, Casilli, F, Aalbers, F, Roelfes, G. | | Deposit date: | 2024-11-01 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | An artificial copper-Michaelase featuring a genetically encoded bipyridine ligand for asymmetric additions to nitroalkenes.
Angew.Chem.Int.Ed.Engl., 2025
|
|
7QZ8
 
 | |
7QZ7
 
 | |
7QZ9
 
 | |
7QZ5
 
 | |
