5L92
 
 | |
5L90
 
 | |
6TB0
 
 | | Crystal structure of thermostable omega transaminase 4-fold mutant from Pseudomonas jessenii | | Descriptor: | Aspartate aminotransferase family protein, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Capra, N, Rozeboom, H.J, Thunnissen, A.M.W.H, Janssen, D.B. | | Deposit date: | 2019-10-31 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Robust omega-Transaminases by Computational Stabilization of the Subunit Interface.
Acs Catalysis, 10, 2020
|
|
3H8G
 
 | | Bestatin complex structure of leucine aminopeptidase from Pseudomonas putida | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, BICARBONATE ION, Cytosol aminopeptidase, ... | | Authors: | Kale, A, Dijkstra, B.W, Sonke, T, Thunnissen, A.M.W.H. | | Deposit date: | 2009-04-29 | | Release date: | 2010-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the leucine aminopeptidase from Pseudomonas putida reveals the molecular basis for its enantioselectivity and broad substrate specificity.
J.Mol.Biol., 398, 2010
|
|
3H8E
 
 | |
6TB1
 
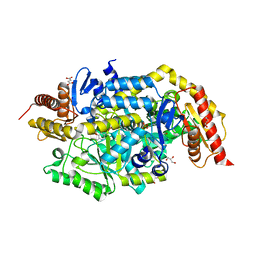 | | Crystal structure of thermostable omega transaminase 6-fold mutant from Pseudomonas jessenii | | Descriptor: | Aspartate aminotransferase family protein, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Capra, N, Rozeboom, H.J, Thunnissen, A.M.W.H, Janssen, D.B. | | Deposit date: | 2019-10-31 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Robust omega-Transaminases by Computational Stabilization of the Subunit Interface.
Acs Catalysis, 10, 2020
|
|
3H8F
 
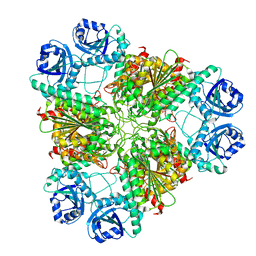 | | High pH native structure of leucine aminopeptidase from Pseudomonas putida | | Descriptor: | BICARBONATE ION, Cytosol aminopeptidase, MANGANESE (II) ION, ... | | Authors: | Kale, A, Dijkstra, B.W, Sonke, T, Thunnissen, A.M.W.H. | | Deposit date: | 2009-04-29 | | Release date: | 2010-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the leucine aminopeptidase from Pseudomonas putida reveals the molecular basis for its enantioselectivity and broad substrate specificity.
J.Mol.Biol., 398, 2010
|
|
5LH9
 
 | |
5LHA
 
 | |
6F8C
 
 | |
6F85
 
 | |
6F8A
 
 | |
6FPS
 
 | |
6FUU
 
 | |
6GK5
 
 | |
6GK6
 
 | |
1OXX
 
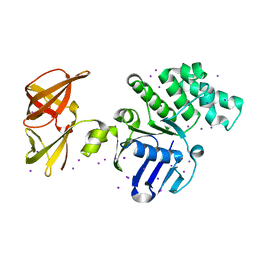 | | Crystal structure of GlcV, the ABC-ATPase of the glucose ABC transporter from Sulfolobus solfataricus | | Descriptor: | ABC transporter, ATP binding protein, IODIDE ION | | Authors: | Verdon, G, Albers, S.-V, van Oosterwijk, N, Dijkstra, B.W, Driessen, A.J.M, Thunnissen, A.M.W.H. | | Deposit date: | 2003-04-03 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Formation of the productive ATP-Mg2+-bound dimer of GlcV, an ABC-ATPase from Sulfolobus solfataricus
J.Mol.Biol., 334, 2003
|
|
4OXV
 
 | |
4P0G
 
 | |
4OZ9
 
 | |
4OWD
 
 | |
4OYV
 
 | |
4P11
 
 | |
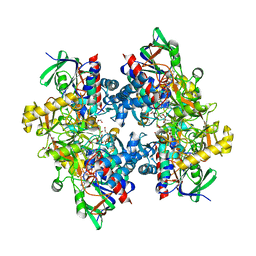
4C3Y
 
 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 1,4-androstadiene-3,17- dione | | Descriptor: | 3-KETOSTEROID DEHYDROGENASE, ANDROSTA-1,4-DIENE-3,17-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rohman, A, van Oosterwijk, N, Thunnissen, A.M.W.H, Dijkstra, B.W. | | Deposit date: | 2013-08-28 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis of 3-Ketosteroid Delta1-Dehydrogenase from Rhodococcus Erythropolis Sq1 Explain its Catalytic Mechanism
J.Biol.Chem., 288, 2013
|
|
4C3X
 
 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 | | Descriptor: | 3-KETOSTEROID DEHYDROGENASE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rohman, A, van Oosterwijk, N, Thunnissen, A.M.W.H, Dijkstra, B.W. | | Deposit date: | 2013-08-28 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis of 3-Ketosteroid Delta1-Dehydrogenase from Rhodococcus Erythropolis Sq1 Explain its Catalytic Mechanism
J.Biol.Chem., 288, 2013
|
|
