1QTE
 
 | | CRYSTAL STRUCTURE OF THE 70 KDA SOLUBLE LYTIC TRANSGLYCOSYLASE SLT70 FROM ESCHERICHIA COLI AT 1.90 A RESOLUTION IN COMPLEX WITH A 1,6-ANHYDROMUROTRIPEPTIDE | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | van Asselt, E.J, Thunnissen, A.-M.W.H, Dijkstra, B.W. | | Deposit date: | 1999-06-27 | | Release date: | 1999-08-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution crystal structures of the Escherichia coli lytic transglycosylase Slt70 and its complex with a peptidoglycan fragment.
J.Mol.Biol., 291, 1999
|
|
4ZZD
 
 | |
8ABS
 
 | | Crystal structure of CYP109A2 from Bacillus megaterium bound with testosterone and putative ligand 4,6-dimethyloctanoic acid | | Descriptor: | (4~{S},6~{S})-4,6-dimethyloctanoic acid, Cytochrome P450, HEME B/C, ... | | Authors: | Jozwik, I.K, Rozeboom, H.J, Thunnissen, A.-M.W.H. | | Deposit date: | 2022-07-04 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Regio- and stereoselective steroid hydroxylation by CYP109A2 from Bacillus megaterium explored by X-ray crystallography and computational modeling.
Febs J., 290, 2023
|
|
8ABR
 
 | | Crystal structure of CYP109A2 from Bacillus megaterium bound with putative ligands hexanoic acid and octanoic acid | | Descriptor: | Cytochrome P450, HEME B/C, HEXANOIC ACID, ... | | Authors: | Jozwik, I.K, Rozeboom, H.J, Thunnissen, A.-M.W.H. | | Deposit date: | 2022-07-04 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Regio- and stereoselective steroid hydroxylation by CYP109A2 from Bacillus megaterium explored by X-ray crystallography and computational modeling.
Febs J., 290, 2023
|
|
1QSA
 
 | | CRYSTAL STRUCTURE OF THE 70 KDA SOLUBLE LYTIC TRANSGLYCOSYLASE SLT70 FROM ESCHERICHIA COLI AT 1.65 ANGSTROMS RESOLUTION | | Descriptor: | ACETATE ION, GLYCEROL, PROTEIN (SOLUBLE LYTIC TRANSGLYCOSYLASE SLT70), ... | | Authors: | van Asselt, E.J, Thunnissen, A.-M.W.H, Dijkstra, B.W. | | Deposit date: | 1999-06-20 | | Release date: | 1999-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High resolution crystal structures of the Escherichia coli lytic transglycosylase Slt70 and its complex with a peptidoglycan fragment.
J.Mol.Biol., 291, 1999
|
|
3T36
 
 | |
4HJV
 
 | | Crystal structure of E. coli MltE with bound bulgecin and murodipeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid, 4-O-(4-O-SULFONYL-N-ACETYLGLUCOSAMININYL)-5-METHYLHYDROXY-L-PROLINE-TAURINE, Endo-type membrane-bound lytic murein transglycosylase A, ... | | Authors: | Fibriansah, G, Gliubich, F.I, Thunnissen, A.-M.W.H. | | Deposit date: | 2012-10-14 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | On the Mechanism of Peptidoglycan Binding and Cleavage by the endo-Specific Lytic Transglycosylase MltE from Escherichia coli.
Biochemistry, 51, 2012
|
|
4HJZ
 
 | | 1.9 A Crystal structure of E. coli MltE-E64Q with bound chitopentaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Fibriansah, G, Gliubich, F.I, Thunnissen, A.-M.W.H. | | Deposit date: | 2012-10-14 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | On the Mechanism of Peptidoglycan Binding and Cleavage by the endo-Specific Lytic Transglycosylase MltE from Escherichia coli.
Biochemistry, 51, 2012
|
|
4HJY
 
 | | 2.4 A Crystal structure of E. coli MltE-E64Q with bound chitopentaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endo-type membrane-bound lytic murein transglycosylase A | | Authors: | Fibriansah, G, Gliubich, F.I, Thunnissen, A.-M.W.H. | | Deposit date: | 2012-10-14 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | On the Mechanism of Peptidoglycan Binding and Cleavage by the endo-Specific Lytic Transglycosylase MltE from Escherichia coli.
Biochemistry, 51, 2012
|
|
3R6Q
 
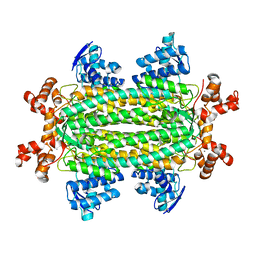 | | A triclinic-lattice structure of aspartase from Bacillus sp. YM55-1 | | Descriptor: | Aspartase, CALCIUM ION | | Authors: | Fibriansah, G, Puthan Veetil, V, Poelarends, G.J, Thunnissen, A.-M.W.H. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the catalytic mechanism of aspartate ammonia lyase.
Biochemistry, 50, 2011
|
|
3R6Y
 
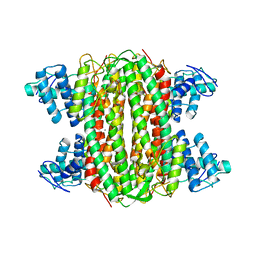 | | Crystal structure of chymotrypsin-treated aspartase from Bacillus sp. YM55-1 | | Descriptor: | Aspartase, CALCIUM ION | | Authors: | Fibriansah, G, Puthan Veetil, V, Poelarends, G.J, Thunnissen, A.-M.W.H. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the catalytic mechanism of aspartate ammonia lyase.
Biochemistry, 50, 2011
|
|
3R6V
 
 | | Crystal structure of aspartase from Bacillus sp. YM55-1 with bound L-aspartate | | Descriptor: | ASPARTIC ACID, Aspartase, CALCIUM ION | | Authors: | Fibriansah, G, Puthan Veetil, V, Poelarends, G.J, Thunnissen, A.-M.W.H. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the catalytic mechanism of aspartate ammonia lyase.
Biochemistry, 50, 2011
|
|
3RYB
 
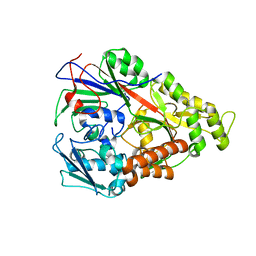 | | Lactococcal OppA complexed with SLSQSLSQS | | Descriptor: | Oligopeptide, Oligopeptide-binding protein oppA | | Authors: | Berntsson, R.P.-A, Thunnissen, A.-M.W.H, Poolman, B, Slotboom, D.-J. | | Deposit date: | 2011-05-11 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Importance of a Hydrophobic Pocket for Peptide Binding in Lactococcal OppA.
J.Bacteriol., 193, 2011
|
|
3RYA
 
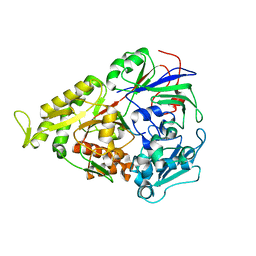 | | Lactococcal OppA complexed with SLSQLSSQS | | Descriptor: | Oligopeptide, Oligopeptide-binding protein oppA | | Authors: | Berntsson, R.P.-A, Thunnissen, A.-M.W.H, Poolman, B, Slotboom, D.-J. | | Deposit date: | 2011-05-11 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Importance of a Hydrophobic Pocket for Peptide Binding in Lactococcal OppA.
J.Bacteriol., 193, 2011
|
|
3F8C
 
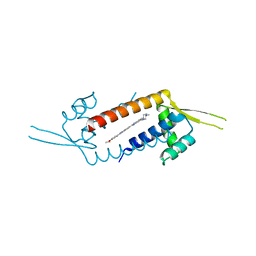 | | Crystal structure of multidrug binding transcriptional regulator LmrR complexed with Hoechst 33342 | | Descriptor: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, Transcriptional regulator, PadR-like family | | Authors: | Madoori, P.K, Agustiandari, H, Driessen, A.J.M, Thunnissen, A.-M.W.H. | | Deposit date: | 2008-11-12 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the transcriptional regulator LmrR and its mechanism of multidrug recognition.
Embo J., 28, 2009
|
|
3F8B
 
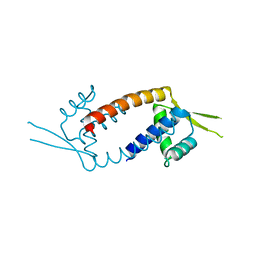 | | Crystal structure of the multidrug binding transcriptional regulator LmrR in drug free state | | Descriptor: | Transcriptional regulator, PadR-like family | | Authors: | Madoori, P.K, Agustiandari, H, Driessen, A.J.M, Thunnissen, A.-M.W.H. | | Deposit date: | 2008-11-12 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the transcriptional regulator LmrR and its mechanism of multidrug recognition.
Embo J., 28, 2009
|
|
3F8F
 
 | | Crystal structure of multidrug binding transcriptional regulator LmrR complexed with Daunomycin | | Descriptor: | DAUNOMYCIN, Transcriptional regulator, PadR-like family | | Authors: | Madoori, P.K, Agustiandari, H, Driessen, A.J.M, Thunnissen, A.-M.W.H. | | Deposit date: | 2008-11-12 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the transcriptional regulator LmrR and its mechanism of multidrug recognition.
Embo J., 28, 2009
|
|
3FTO
 
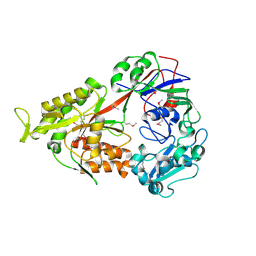 | | Crystal structure of OppA in a open conformation | | Descriptor: | Oligopeptide-binding protein oppA | | Authors: | Berntsson, R.P.-A, Oktaviani, N.A, Fusetti, F, Thunnissen, A.-M.W.H, Poolman, B, Slotboom, D.-J. | | Deposit date: | 2009-01-13 | | Release date: | 2009-03-31 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Selenomethionine incorporation in proteins expressed in Lactococcus lactis.
Protein Sci., 18, 2009
|
|
