6D2V
 
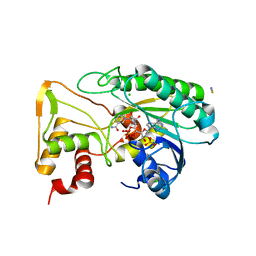 | | Apo Structure of TerB, an NADP Dependent Oxidoreductase in the Terfestatin Biosynthesis Pathway | | 分子名称: | CHLORIDE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, THIOCYANATE ION, ... | | 著者 | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | 登録日 | 2018-04-14 | | 公開日 | 2018-06-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
8QDU
 
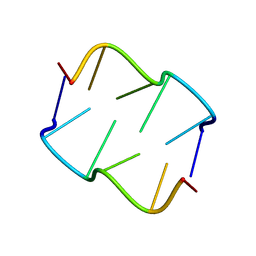 | | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions | | 分子名称: | DNA (5'-D(*(FC)P*GP*(FC)P*GP*(FC)P*G)-3') | | 著者 | El-Khoury, R, Cabrero, C, Movilla, S, Thorpe, J.D, Roman, M, Orozco, M, Gonzalez, C, Damha, M.J. | | 登録日 | 2023-08-30 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-31 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions.
Nucleic Acids Res., 52, 2024
|
|
6WF4
 
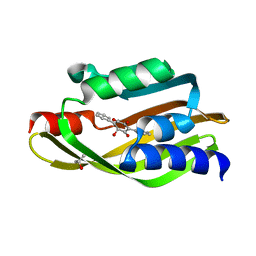 | | Crystal Structure of TerC Co-crystallized with Polyporic Acid | | 分子名称: | (2~5~S)-2~3~,2~5~,2~6~-trihydroxy[1~1~,2~1~:2~4~,3~1~-terphenyl]-2~2~(2~5~H)-one, ISOPROPYL ALCOHOL, Terfestatin Biosyntheis Enzyme C | | 著者 | Clinger, J.A, Miller, M.D, Hall, R.E, Zhang, Y, Elshahawi, S.I, Thorson, J.S, Van Lanen, S.G, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2020-04-03 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structural and functional characterization of two cooperative enzymes responsible for the stability
of p-terphenyls.
To be published
|
|
6UBL
 
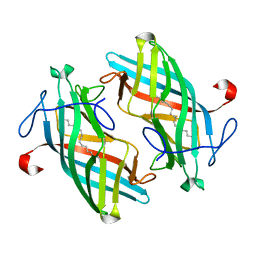 | | Structure of DynF from the Dynemicin Biosynthesis Pathway of Micromonospora chersina | | 分子名称: | DynF, PALMITIC ACID | | 著者 | Kosgei, A.J, Miller, M.D, Xu, W, Bhardwaj, M, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | 登録日 | 2019-09-12 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.499 Å) | | 主引用文献 | The crystal structure of DynF from the dynemicin-biosynthesis pathway of Micromonospora chersina.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
6VQP
 
 | | Structure of CalU17 from the Calicheamicin Biosynthesis Pathway of Micromonospora echinospora | | 分子名称: | CalU17, CalU17 His-Tagged protein, GLYCEROL, ... | | 著者 | Kosgei, A.J, Miller, M.D, Xu, W, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | 登録日 | 2020-02-05 | | 公開日 | 2021-02-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of DynF from the Dynemicin Biosynthesis Pathway of Micromonospora chersina
To Be Published
|
|
6D34
 
 | | Apo Crystal Structure of TerC, a Terfestatin Biosynthesis Enzyme | | 分子名称: | ISOPROPYL ALCOHOL, TerC | | 著者 | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | 登録日 | 2018-04-14 | | 公開日 | 2018-06-13 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
8QE4
 
 | | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions | | 分子名称: | DNA (5'-D(*CP*(FRG)P*CP*(FRG)P*CP*(FRG))-3') | | 著者 | El-Khoury, R, Cabrero, C, Movilla, S, Friedland, D, Thorpe, J.D, Roman, M, Orozco, M, Gonzalez, C, Damha, M. | | 登録日 | 2023-08-30 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-31 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions.
Nucleic Acids Res., 52, 2024
|
|
4X8G
 
 | | Crystal structure of human peptidylarginine deiminase type4 (PAD4) in complex with GSK199 | | 分子名称: | CALCIUM ION, Protein-arginine deiminase type-4, [(3R)-3-aminopiperidin-1-yl][2-(1-ethyl-1H-pyrrolo[2,3-b]pyridin-2-yl)-7-methoxy-1-methyl-1H-benzimidazol-5-yl]methanone | | 著者 | Lewis, H.D, Bax, B.D, Chung, C.-W, Polyakova, O, Thorpe, J. | | 登録日 | 2014-12-10 | | 公開日 | 2015-01-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.29 Å) | | 主引用文献 | Inhibition of PAD4 activity is sufficient to disrupt mouse and human NET formation.
Nat.Chem.Biol., 11, 2015
|
|
5EEG
 
 | | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with tetrazole-SAH | | 分子名称: | (2~{R},3~{R},4~{S},5~{S})-2-(6-aminopurin-9-yl)-5-[[(3~{S})-3-azanyl-3-(1~{H}-1,2,3,4-tetrazol-5-yl)propyl]sulfanylmethyl]oxolane-3,4-diol, Carminomycin 4-O-methyltransferase DnrK | | 著者 | Wang, F, Singh, S, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-10-22 | | 公開日 | 2015-12-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.255 Å) | | 主引用文献 | Functional AdoMet Isosteres Resistant to Classical AdoMet Degradation Pathways.
Acs Chem.Biol., 11, 2016
|
|
6UK5
 
 | | Structure of SAM bound CalS10, an amino pentose methyltransferase from Micromonospora echinaspora involved in calicheamicin biosynthesis | | 分子名称: | ACETATE ION, CalS10, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Alvarado, S.K, Miller, M.D, Xu, W, Wang, Z, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | 登録日 | 2019-10-04 | | 公開日 | 2020-10-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of SAM bound CalS10, an amino pentose methyltransferase from Micromonospora echinaspora involved in calicheamicin biosynthesis
To Be Published
|
|
5EEH
 
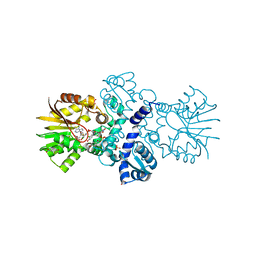 | | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with SAH and 2-chloro-4-nitrophenol | | 分子名称: | 2-chloranyl-4-nitro-phenol, Carminomycin 4-O-methyltransferase DnrK, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Wang, F, Singh, S, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-10-22 | | 公開日 | 2015-12-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Functional AdoMet Isosteres Resistant to Classical AdoMet Degradation Pathways.
Acs Chem.Biol., 11, 2016
|
|
4WS9
 
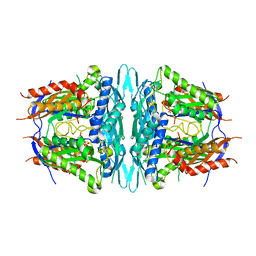 | | Crystal structure of sMAT N159G from Sulfolobus solfataricus | | 分子名称: | PHOSPHATE ION, S-adenosylmethionine synthase | | 著者 | Wang, F, Brady, E.L, Singh, S, Clinger, J.A, Huber, T.D, Thorson, J.S, Phillips Jr, G.N. | | 登録日 | 2014-10-26 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.803 Å) | | 主引用文献 | Crystal structure of sMAT N159G from Sulfolobus solfataricus.
To Be Published
|
|
4X8C
 
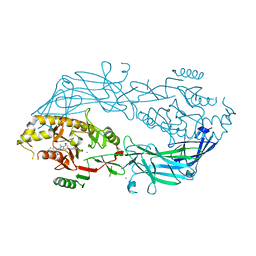 | | Crystal structure of human peptidylarginine deiminase type4 (PAD4) in complex with GSK147 | | 分子名称: | CALCIUM ION, Protein-arginine deiminase type-4, [(3S,4R)-3-amino-4-hydroxypiperidin-1-yl]{2-[1-(cyclopropylmethyl)-1H-pyrrolo[2,3-b]pyridin-2-yl]-7-methoxy-1-methyl-1H-benzimidazol-5-yl}methanone | | 著者 | Lewis, H.D, Bax, B.D, Chung, C.-W, Polyakova, O, Thorpe, J. | | 登録日 | 2014-12-10 | | 公開日 | 2015-01-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Inhibition of PAD4 activity is sufficient to disrupt mouse and human NET formation.
Nat.Chem.Biol., 11, 2015
|
|
1YOW
 
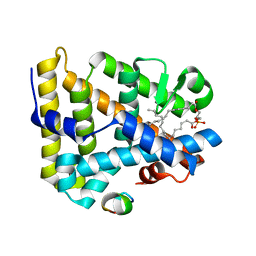 | | human Steroidogenic Factor 1 LBD with bound Co-factor Peptide | | 分子名称: | PHOSPHATIDYL ETHANOL, Steroidogenic factor 1, TIF2 peptide | | 著者 | Krylova, I.N, Sablin, E.P, Xu, R.X, Waitt, G.M, Juzumiene, D, Williams, J.D, Ingraham, H.A, Willson, T.M, Williams, S.P, Montana, V, Madauss, K.P, Moore, J, Bynum, J.M, Lebedeva, L, MacKay, J.A, Suzawa, M, Guy, R.K, Thornton, J.W. | | 登録日 | 2005-01-28 | | 公開日 | 2005-05-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1
Cell(Cambridge,Mass.), 120, 2005
|
|
1YOK
 
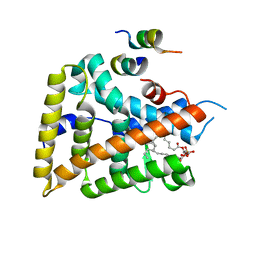 | | crystal structure of human LRH-1 bound with TIF-2 peptide and phosphatidylglycerol | | 分子名称: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Nuclear receptor coactivator 2, Orphan nuclear receptor NR5A2 | | 著者 | Krylova, I.N, Sablin, E.P, Moore, J, Xu, R.X, Waitt, G.M, MacKay, J.A, Juzumiene, D, Bynum, J.M, Madauss, K, Montana, V, Lebedeva, L, Suzawa, M, Williams, J.D, Williams, S.P, Guy, R.K, Thornton, J.W, Fletterick, R.J, Willson, T.M, Ingraham, H.A. | | 登録日 | 2005-01-27 | | 公開日 | 2005-07-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1.
Cell(Cambridge,Mass.), 120, 2005
|
|
6UV6
 
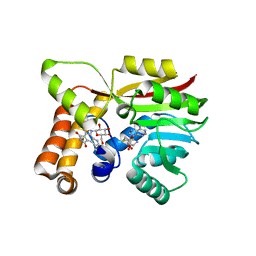 | | AtmM with bound rebeccamycin analogue | | 分子名称: | 12-beta-D-glucopyranosyl-12,13-dihydro-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazole-5,7(6H)-dione, D-glucose O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Alvarado, S.K, Wang, Z, Miller, M.D, Thorson, J.S, Phillips Jr, G.N. | | 登録日 | 2019-11-01 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | Structure of AtmM Bound with Glycosylated Indolocarbazole
To Be Published
|
|
5K9M
 
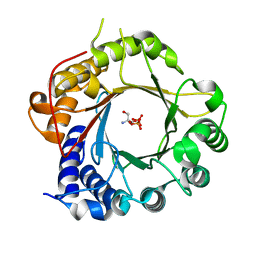 | | Crystal Structure of PriB Binary Complex with Product Diphosphate | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PYROPHOSPHATE 2-, PriB Prenyltransferase | | 著者 | Cao, H, Elshahawi, S, Benach, J, Wasserman, S.R, Morisco, L.L, Koss, J.W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2016-06-01 | | 公開日 | 2016-06-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure and specificity of a permissive bacterial C-prenyltransferase.
Nat. Chem. Biol., 13, 2017
|
|
5JXM
 
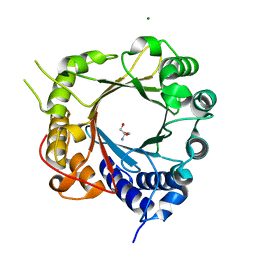 | | Crystal Structure of Prenyltransferase PriB Apo Form | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, PriB | | 著者 | Cao, H, Elshahawi, S, Benach, J, Wasserman, S.R, Morisco, L.L, Koss, J.W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2016-05-13 | | 公開日 | 2016-06-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Structure and specificity of a permissive bacterial C-prenyltransferase.
Nat. Chem. Biol., 13, 2017
|
|
5INJ
 
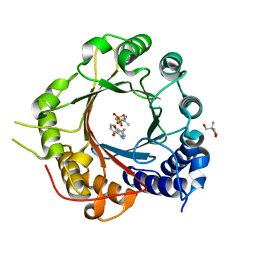 | | Crystal Structure of Prenyltransferase PriB Ternary Complex with L-Tryptophan and Dimethylallyl thiolodiphosphate (DMSPP) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Prenyltransferase, ... | | 著者 | Cao, H, Elshahawi, S, Benach, J, Wasserman, S.R, Morisco, L.L, Koss, J.W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2016-03-07 | | 公開日 | 2016-05-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure and specificity of a permissive bacterial C-prenyltransferase.
Nat. Chem. Biol., 13, 2017
|
|
3TOS
 
 | | Crystal Structure of CalS11, Calicheamicin Methyltransferase | | 分子名称: | 1,2-ETHANEDIOL, CalS11, GLUTAMIC ACID, ... | | 著者 | Chang, A, Aceti, D.J, Beebe, E.T, Makino, S.-I, Wrobel, R.L, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2011-09-06 | | 公開日 | 2011-10-05 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal Structure of CalS11, Calicheamicin methyltransferase
To be Published
|
|
1QOK
 
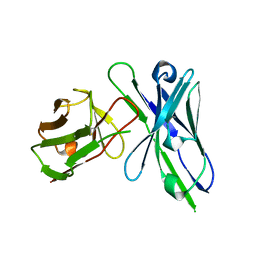 | | MFE-23 AN ANTI-CARCINOEMBRYONIC ANTIGEN SINGLE-CHAIN FV ANTIBODY | | 分子名称: | MFE-23 RECOMBINANT ANTIBODY FRAGMENT | | 著者 | Boehm, M.K, Corper, A.L, Wan, T, Sohi, M.K, Sutton, B.J, Thornton, J.D, Keep, P.A, Chester, K.A, Begent, R.H.J, Perkins, S.J. | | 登録日 | 1999-11-11 | | 公開日 | 2000-11-10 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of the Anti-Carcinoembryonic Antigen Single-Chain Fv Antibody Mfe-23 and a Model for Antigen Binding Based on Intermolecular Contacts
Biochem.J., 346, 2000
|
|
4M83
 
 | | Ensemble refinement of protein crystal structure (2IYF) of macrolide glycosyltransferases OleD complexed with UDP and Erythromycin A | | 分子名称: | ERYTHROMYCIN A, MAGNESIUM ION, Oleandomycin glycosyltransferase, ... | | 著者 | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2013-08-12 | | 公開日 | 2013-09-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.698 Å) | | 主引用文献 | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
1ZNM
 
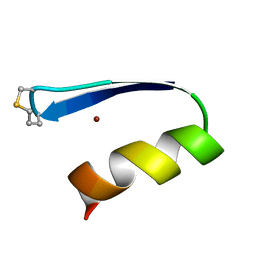 | | A zinc finger with an artificial beta-turn, original sequence taken from the third zinc finger domain of the human transcriptional repressor protein YY1 (YING and YANG 1, a delta transcription factor), nmr, 34 structures | | 分子名称: | YY1, ZINC ION | | 著者 | Viles, J.H, Patel, S.U, Mitchell, J.B.O, Moody, C.M, Justice, D.E, Uppenbrink, J, Doyle, P.M, Harris, C.J, Sadler, P.J, Thornton, J.M. | | 登録日 | 1997-11-20 | | 公開日 | 1998-04-01 | | 最終更新日 | 2017-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design, synthesis and structure of a zinc finger with an artificial beta-turn.
J.Mol.Biol., 279, 1998
|
|
1MP5
 
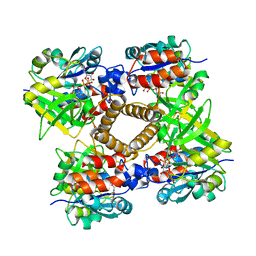 | | Y177F VARIANT OF S. ENTERICA RmlA | | 分子名称: | URIDINE-5'-DIPHOSPHATE-GLUCOSE, Y177F VARIANT OF S. ENTERICA RmlA BOUND TO UDP-GLUCOSE | | 著者 | Barton, W.A, Biggins, J.B, Jiang, J, Thorson, J.S, Nikolov, D.B. | | 登録日 | 2002-09-11 | | 公開日 | 2002-10-09 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Expanding pyrimidine diphosphosugar libraries via structure-based nucleotidylyltransferase engineering
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1MP4
 
 | | W224H VARIANT OF S. ENTERICA RmlA | | 分子名称: | URIDINE-5'-DIPHOSPHATE-GLUCOSE, W224H Variant of S. Enterica RmlA Bound to UDP-Glucose | | 著者 | Barton, W.A, Biggins, J.B, Jiang, J, Thorson, J.S, Nikolov, D.B. | | 登録日 | 2002-09-11 | | 公開日 | 2002-10-09 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Expanding pyrimidine diphosphosugar libraries via structure-based nucleotidylyltransferase engineering
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
