3HVM
 
 | |
8EYZ
 
 | | Engineered glutamine binding protein bound to GLN and a cobaloxime ligand | | Descriptor: | AZIDOBIS (DIMETHYLGLYOXIMATO) PYRIDINECOBALT, Amino acid ABC transporter substrate-binding protein, GLUTAMINE, ... | | Authors: | Bridwell-Rabb, J, Boggs, D.G, Olshansky, L, Fatima, S, Thompson, P. | | Deposit date: | 2022-10-29 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Engineering a Conformationally Switchable Artificial Metalloprotein.
J.Am.Chem.Soc., 144, 2022
|
|
1XGK
 
 | | CRYSTAL STRUCTURE OF N12G AND A18G MUTANT NMRA | | Descriptor: | CHLORIDE ION, GLYCEROL, NITROGEN METABOLITE REPRESSION REGULATOR NMRA, ... | | Authors: | Lamb, H.K, Ren, J, Park, A, Johnson, C, Leslie, K, Cocklin, S, Thompson, P, Mee, C, Cooper, A, Stammers, D.K, Hawkins, A.R. | | Deposit date: | 2004-09-17 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Modulation of the ligand binding properties of the transcription repressor NmrA by GATA-containing DNA and site-directed mutagenesis
Protein Sci., 13, 2004
|
|
1TI7
 
 | | CRYSTAL STRUCTURE OF NMRA, A NEGATIVE TRANSCRIPTIONAL REGULATOR, IN COMPLEX WITH NADP AT 1.7A RESOLUTION | | Descriptor: | CHLORIDE ION, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lamb, H.K, Leslie, K, Dodds, A.L, Nutley, M, Cooper, A, Johnson, C, Thompson, P, Stammers, D.K, Hawkins, A.R. | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The negative transcriptional regulator NmrA discriminates between oxidized and reduced dinucleotides.
J.Biol.Chem., 278, 2003
|
|
4UFC
 
 | | Crystal structure of the GH95 enzyme BACOVA_03438 | | Descriptor: | CACODYLATE ION, CALCIUM ION, GH95, ... | | Authors: | Rogowski, A, Briggs, J.A, Mortimer, J.C, Tryfona, T, Terrapon, N, Lowe, E.C, Basle, A, Morland, C, Day, A.M, Zheng, H, Rogers, T.E, Thompson, P, Hawkins, A.R, Yadav, M.P, Henrissat, B, Martens, E.C, Dupree, P, Gilbert, H.J, Bolam, D.N. | | Deposit date: | 2015-03-16 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Glycan Complexity Dictates Microbial Resource Allocation in the Large Intestine.
Nat.Commun., 6, 2015
|
|
2ZNM
 
 | | Oxidoreductase NmDsbA3 from Neisseria meningitidis | | Descriptor: | Thiol:disulfide interchange protein DsbA | | Authors: | Vivian, J.P, Scoullar, J, Robertson, A.L, Bottomley, S.P, Horne, J, Chin, Y, Velkov, T, Wielens, J, Thompson, P.E, Piek, S, Byres, E, Beddoe, T, Wilce, M.C.J, Kahler, C, Rossjohn, J, Scanlon, M.J. | | Deposit date: | 2008-04-30 | | Release date: | 2008-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Characterization of the Oxidoreductase NmDsbA3 from Neisseria meningitidis
J.Biol.Chem., 283, 2008
|
|
3B1U
 
 | | Crystal structure of human peptidylarginine deiminase 4 in complex with o-F-amidine | | Descriptor: | 2-{[(2S)-1-amino-5-{[(1Z)-2-fluoroethanimidoyl]amino}-1-oxopentan-2-yl]carbamoyl}benzoic acid, CALCIUM ION, Protein-arginine deiminase type-4, ... | | Authors: | Causey, C.P, Jones, J.E, Slack, J.L, Kamei, D, Jones Jr, L.E, Subramanian, V, Knuckley, B, Ebrahimi, P, Chumanevich, A.A, Luo, Y, Hashimoto, H, Shimizu, T, Sato, M, Hofseth, L.J, Thompson, P.R. | | Deposit date: | 2011-07-13 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Development of N-alpha-(2-Carboxyl)benzoyl-N(5)-(2-fluoro-1-iminoethyl)-l-ornithine Amide (o-F-amidine) and N-alpha-(2-Carboxyl)benzoyl-N(5)-(2-chloro-1-iminoethyl)-l-ornithine Amide (o-Cl-amidine) As Second Generation Protein Arginine Deiminase (PAD) Inhibitors
J.Med.Chem., 54, 2011
|
|
4UIO
 
 | | Structure of the Salmonella typhi Type I Dehydroquinase covalently inhibited by a 3-dehydroquinic acid derivative | | Descriptor: | (1~{R},3~{R},4~{S},5~{R})-3-methyl-1,3,4,5-tetrakis(oxidanyl)cyclohexane-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, CHLORIDE ION, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Tizon, L, Lence, E, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Chemical Modification of a Dehydratase Enzyme Involved in Bacterial Virulence by an Ammonium Derivative: Evidence of its Active Site Covalent Adduct.
J.Am.Chem.Soc., 137, 2015
|
|
3JBJ
 
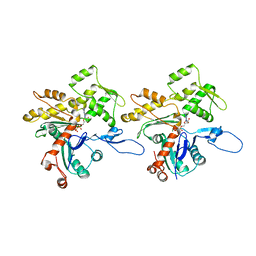 | | Cryo-EM reconstruction of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Kim, L.Y, Thompson, P.M, Lee, H.T, Pershad, M, Campbell, S.L, Alushin, G.M. | | Deposit date: | 2015-09-03 | | Release date: | 2015-11-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | The Structural Basis of Actin Organization by Vinculin and Metavinculin.
J.Mol.Biol., 428, 2016
|
|
3JBK
 
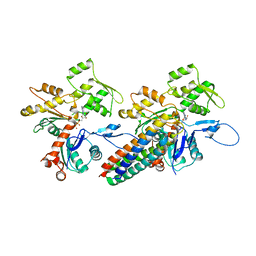 | | Cryo-EM reconstruction of the metavinculin-actin interface | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Kim, L.Y, Thompson, P.M, Lee, H.T, Pershad, M, Campbell, S.L, Alushin, G.M. | | Deposit date: | 2015-09-03 | | Release date: | 2015-11-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | The Structural Basis of Actin Organization by Vinculin and Metavinculin.
J.Mol.Biol., 428, 2016
|
|
3JBI
 
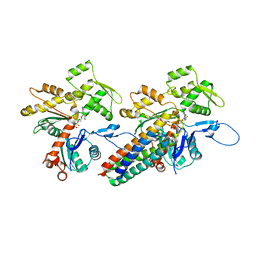 | | MDFF model of the vinculin tail domain bound to F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Kim, L.Y, Thompson, P.M, Lee, H.T, Pershad, M, Campbell, S.L, Alushin, G.M. | | Deposit date: | 2015-09-02 | | Release date: | 2015-11-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | The Structural Basis of Actin Organization by Vinculin and Metavinculin.
J.Mol.Biol., 428, 2016
|
|
1EBF
 
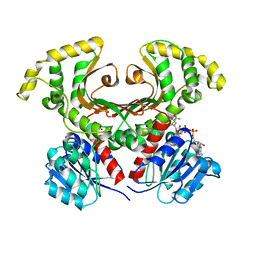 | | HOMOSERINE DEHYDROGENASE FROM S. CEREVISIAE COMPLEX WITH NAD+ | | Descriptor: | HOMOSERINE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | DeLaBarre, B, Thompson, P.R, Wright, G.D, Berghuis, A.M. | | Deposit date: | 2000-01-24 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of homoserine dehydrogenase suggest a novel catalytic mechanism for oxidoreductases.
Nat.Struct.Biol., 7, 2000
|
|
1EBU
 
 | | HOMOSERINE DEHYDROGENASE COMPLEX WITH NAD ANALOGUE AND L-HOMOSERINE | | Descriptor: | 3-AMINOMETHYL-PYRIDINIUM-ADENINE-DINUCLEOTIDE, HOMOSERINE DEHYDROGENASE, L-HOMOSERINE, ... | | Authors: | DeLaBarre, B, Thompson, P.R, Wright, G.D, Berghuis, A.M. | | Deposit date: | 2000-01-24 | | Release date: | 2000-03-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of homoserine dehydrogenase suggest a novel catalytic mechanism for oxidoreductases.
Nat.Struct.Biol., 7, 2000
|
|
3ZEU
 
 | | Structure of a Salmonella typhimurium YgjD-YeaZ heterodimer bound to ATPgammaS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Nichols, C.E, Lamb, H.K, Thompson, P, El Omari, K, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-20 | | Last modified: | 2017-03-29 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Crystal Structure of the Dimer of Two Essential Salmonella Typhimurium Proteins, Ygjd & Yeaz and Calorimetric Evidence for the Formation of a Ternary Ygjd-Yeaz-Yjee Complex.
Protein Sci., 22, 2013
|
|
3ZET
 
 | | Structure of a Salmonella typhimurium YgjD-YeaZ heterodimer. | | Descriptor: | ADENOSINE MONOPHOSPHATE, CADMIUM ION, PROBABLE TRNA THREONYLCARBAMOYLADENOSINE BIOSYNTHESIS PROTEIN GCP, ... | | Authors: | Nichols, C.E, Lamb, H.K, Thompson, P, El Omari, K, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-20 | | Last modified: | 2013-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of the Dimer of Two Essential Salmonella Typhimurium Proteins, Ygjd & Yeaz and Calorimetric Evidence for the Formation of a Ternary Ygjd-Yeaz-Yjee Complex.
Protein Sci., 22, 2013
|
|
3Q8M
 
 | | Crystal Structure of Human Flap Endonuclease FEN1 (D181A) in complex with substrate 5'-flap DNA and K+ | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*AP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*T)-3'), ... | | Authors: | Tsutakawa, S.E, Classen, S, Chapados, B.R, Arvai, A, Finger, D.L, Guenther, G, Tomlinson, C.G, Thompson, P, Sarker, A.H, Shen, B, Cooper, P.K, Grasby, J.A, Tainer, J.A. | | Deposit date: | 2011-01-06 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human Flap Endonuclease Structures, DNA Double-Base Flipping, and a Unified Understanding of the FEN1 Superfamily.
Cell(Cambridge,Mass.), 145, 2011
|
|
6H5D
 
 | | Crystal structure of DHQ1 from Salmonella typhi covalently modified by ligand 2 | | Descriptor: | (1~{S},3~{S},4~{R},5~{R})-3-methyl-1,4,5-tris(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Maneiro, M, Lence, E, Otero, J.M, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hydroxylammonium Derivatives for Selective Active-site Lysine Modification in the Anti-virulence Bacterial Target DHQ1 Enzyme.
Org Chem Front, 9, 2019
|
|
6H5G
 
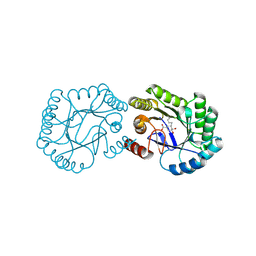 | | Crystal structure of DHQ1 from Salmonella typhi covalently modified by ligand 3 | | Descriptor: | (1~{R},3~{S},4~{R},5~{R})-3-methyl-4,5-bis(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Maneiro, M, Lence, E, Otero, J.M, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Hydroxylammonium Derivatives for Selective Active-site Lysine Modification in the Anti-virulence Bacterial Target DHQ1 Enzyme.
Org Chem Front, 6, 2019
|
|
6H5C
 
 | | Crystal structure of DHQ1 from Salmonella typhi covalently modified by ligand 1 | | Descriptor: | (1~{S},3~{R},4~{S},5~{R})-3-methyl-3,4,5-tris(hydroxyl)cyclohexane-1-carboxylic Acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Maneiro, M, Lence, E, Otero, J.M, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Hydroxylammonium Derivatives for Selective Active-site Lysine Modification in the Anti-virulence Bacterial Target DHQ1 Enzyme.
Org Chem Front, 9, 2019
|
|
3Q8K
 
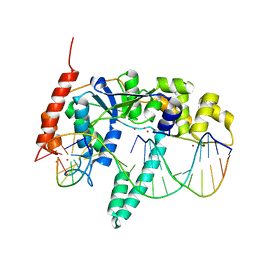 | | Crystal Structure of Human Flap Endonuclease FEN1 (WT) in complex with product 5'-flap DNA, SM3+, and K+ | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*AP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*T)-3'), DNA (5'-D(P*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*T)-3'), ... | | Authors: | Tsutakawa, S.E, Classen, S, Chapados, B.R, Arvai, A, Finger, D.L, Guenther, G, Tomlinson, C.G, Thompson, P, Sarker, A.H, Shen, B, Cooper, P.K, Grasby, J.A, Tainer, J.A. | | Deposit date: | 2011-01-06 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2001 Å) | | Cite: | Human Flap Endonuclease Structures, DNA Double-Base Flipping, and a Unified Understanding of the FEN1 Superfamily.
Cell(Cambridge,Mass.), 145, 2011
|
|
6H5J
 
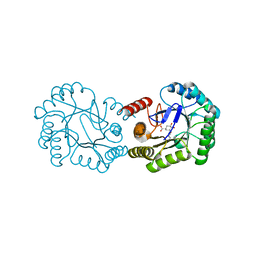 | | Crystal structure of DHQ1 from Salmonella typhi covalently modified by ligand 4 | | Descriptor: | (3~{R})-3,4,5-tris(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Otero, J.M, Maneiro, M, Lence, E, Sanz-Gaitero, M, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Hydroxylammonium Derivatives for Selective Active-site Lysine Modification in the Anti-virulence Bacterial Target DHQ1 Enzyme.
Org Chem Front, 6, 2019
|
|
3Q8L
 
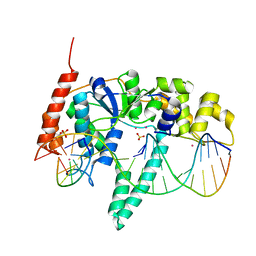 | | Crystal Structure of Human Flap Endonuclease FEN1 (WT) in complex with substrate 5'-flap DNA, SM3+, and K+ | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*AP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*T)-3'), ... | | Authors: | Tsutakawa, S.E, Classen, S, Chapados, B.R, Arvai, A, Finger, D.L, Guenther, G, Tomlinson, C.G, Thompson, P, Sarker, A.H, Shen, B, Cooper, P.K, Grasby, J.A, Tainer, J.A. | | Deposit date: | 2011-01-06 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.319 Å) | | Cite: | Human Flap Endonuclease Structures, DNA Double-Base Flipping, and a Unified Understanding of the FEN1 Superfamily.
Cell(Cambridge,Mass.), 145, 2011
|
|
3DKS
 
 | | DsbA substrate complex | | Descriptor: | Thiol:disulfide interchange protein dsbA, siga peptide | | Authors: | Paxman, J.J, Borg, N.A, Horne, J, Rossjohn, J, Thompson, P.E, Piek, S, Kahler, C.M, Sakellaris, H, Scanlon, M.J. | | Deposit date: | 2008-06-25 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of the bacterial oxidoreductase enzyme DsbA in complex with a peptide reveals a basis for substrate specificity in the catalytic cycle of DsbA enzymes
J.Biol.Chem., 284, 2009
|
|
2LHW
 
 | | Tri-O-GalNAc glycosylated Mucin sequence based on MUC2 Mucin glycoprotein tandem repeat | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MUC2 Mucin Domain Peptide | | Authors: | Borgert, A, Heimburg-Molinaro, J, Lasanajak, Y, Ju, T, Liu, M, Thompson, P, Ragupathi, G, Barany, G, Cummings, R, Smith, D, Live, D. | | Deposit date: | 2011-08-18 | | Release date: | 2012-04-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Deciphering structural elements of mucin glycoprotein recognition.
Acs Chem.Biol., 7, 2012
|
|
2LI2
 
 | | Mono-O-GalNAc glycosylated Mucin sequence based on MUC2 Mucin glycoprotein tandem repeat | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MUC2 Mucin Domain Peptide | | Authors: | Borgert, A, Heimburg-Molinaro, J, Lasanajak, Y, Ju, T, Liu, M, Thompson, P, Ragupathi, G, Barany, G, Cummings, R, Smith, D, Live, D. | | Deposit date: | 2011-08-18 | | Release date: | 2012-04-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Deciphering structural elements of mucin glycoprotein recognition.
Acs Chem.Biol., 7, 2012
|
|
