1W3N
 
 | | Sulfolobus solfataricus 2-keto-3-deoxygluconate (KDG) aldolase complex with D-KDG | | Descriptor: | 2-KETO-3-DEOXY GLUCONATE ALDOLASE, 3-DEOXY-D-ARABINO-HEXONIC ACID, GLYCEROL | | Authors: | Theodossis, A, Walden, H, Westwick, E.J, Connaris, H, Lamble, H.J, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2004-07-17 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural basis for substrate promiscuity in 2-keto-3-deoxygluconate aldolase from the Entner-Doudoroff pathway in Sulfolobus solfataricus.
J. Biol. Chem., 279, 2004
|
|
1W37
 
 | | 2-keto-3-deoxygluconate(KDG) aldolase of Sulfolobus solfataricus | | Descriptor: | 2-KETO-3-DEOXY GLUCONATE ALDOLASE, GLYCEROL, SODIUM ION | | Authors: | Theodossis, A, Walden, H, Westwick, E.J, Connaris, H, Lamble, H.J, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2004-07-13 | | Release date: | 2004-09-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis for substrate promiscuity in 2-keto-3-deoxygluconate aldolase from the Entner-Doudoroff pathway in Sulfolobus solfataricus.
J. Biol. Chem., 279, 2004
|
|
2BKY
 
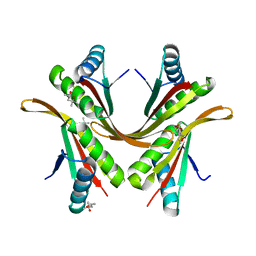 | | Crystal structure of the Alba1:Alba2 heterodimer from sulfolobus solfataricus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA/RNA-BINDING PROTEIN ALBA 1, DNA/RNA-BINDING PROTEIN ALBA 2 | | Authors: | Jelinska, C, Conroy, M.J, Craven, C.J, Bullough, P.A, Waltho, J.P, Taylor, G.L, White, M.F. | | Deposit date: | 2005-02-22 | | Release date: | 2005-07-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Obligate Heterodimerization of the Archaeal Alba2 Protein with Alba1 Provides a Mechanism for Control of DNA Packaging.
Structure, 13, 2005
|
|
2CD9
 
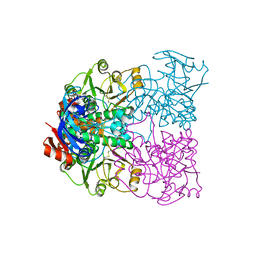 | | Sulfolobus solfataricus Glucose Dehydrogenase 1 - apo form | | Descriptor: | GLUCOSE DEHYDROGENASE, ZINC ION | | Authors: | Milburn, C.C, Lamble, H.J, Theodossis, A, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structural Basis of Substrate Promiscuity in Glucose Dehydrogenase from the Hyperthermophilic Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 281, 2006
|
|
2CDB
 
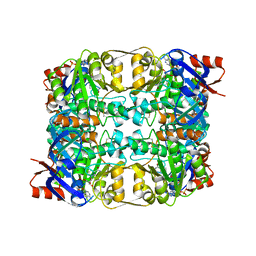 | | Sulfolobus solfataricus Glucose Dehydrogenase 1 in complex with NADP and glucose | | Descriptor: | 1,2-ETHANEDIOL, GLUCOSE 1-DEHYDROGENASE (DHG-1), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Milburn, C.C, Lamble, H.J, Theodossis, A, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structural Basis of Substrate Promiscuity in Glucose Dehydrogenase from the Hyperthermophilic Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 281, 2006
|
|
2CDC
 
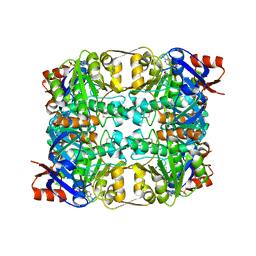 | | Sulfolobus solfataricus Glucose Dehydrogenase 1 in complex with NADP and Xylose | | Descriptor: | 1,2-ETHANEDIOL, GLUCOSE DEHYDROGENASE GLUCOSE 1-DEHYDROGENASE, DHG-1, ... | | Authors: | Milburn, C.C, Lamble, H.J, Theodossis, A, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structural basis of substrate promiscuity in glucose dehydrogenase from the hyperthermophilic archaeon Sulfolobus solfataricus.
J. Biol. Chem., 281, 2006
|
|
2CKJ
 
 | | Human milk xanthine oxidoreductase | | Descriptor: | ACETIC ACID, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Pearson, A.R, Godber, B.L.J, Eisenthal, R, Taylor, G.L, Harrison, R. | | Deposit date: | 2006-04-19 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Human Milk Xanthine Dehydrogenase is Incompletely Converted to the Oxidase Form in the Absence of Proteolysis. A Structural Explanation.
To be Published
|
|
1WCQ
 
 | | Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, SIALIDASE, ... | | Authors: | Newstead, S, Watson, J.N, Bennet, A.J, Taylor, G. | | Deposit date: | 2004-11-19 | | Release date: | 2005-10-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two Nucleophilic Mutants of the Micromonospora Viridifaciens Sialidase Operate with Retention of Configuration by Two Different Mechanisms.
Chembiochem, 6, 2005
|
|
2CDA
 
 | | Sulfolobus solfataricus Glucose Dehydrogenase 1 in complex with NADP | | Descriptor: | GLUCOSE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Milburn, C.C, Lamble, H.J, Theodossis, A, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The Structural Basis of Substrate Promiscuity in Glucose Dehydrogenase from the Hyperthermophilic Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 281, 2006
|
|
2VK5
 
 | | THE STRUCTURE OF CLOSTRIDIUM PERFRINGENS NANI SIALIDASE AND ITS CATALYTIC INTERMEDIATES | | Descriptor: | CALCIUM ION, EXO-ALPHA-SIALIDASE, GLYCEROL | | Authors: | Newstead, S.L, Potter, J.A, Wilson, J.C, Xu, G, Chien, C.H, Watts, A.G, Withers, S.G, Taylor, G.L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | The Structure of Clostridium Perfringens Nani Sialidase and its Catalytic Intermediates.
J.Biol.Chem., 283, 2008
|
|
1ZSQ
 
 | | Crystal Structure of MTMR2 in complex with phosphatidylinositol 3-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-(BUTANOYLOXY)-1-{[(HYDROXY{[2,3,4,6-TETRAHYDROXY-5-(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL)OXY]METHYL}ETHYL BUTANOATE, Myotubularin-related protein 2 | | Authors: | Begley, M.J, Taylor, G.S, Brock, M.A, Ghosh, P, Woods, V.L, Dixon, J.E. | | Deposit date: | 2005-05-25 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Molecular basis for substrate recognition by MTMR2, a myotubularin family phosphoinositide phosphatase
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4GAG
 
 | | Structure of the broadly neutralizing antibody AP33 in complex with its HCV epitope (E2 residues 412-423) | | Descriptor: | GLYCEROL, Genome polyprotein, NEUTRALIZING ANTIBODY AP33 HEAVY CHAIN, ... | | Authors: | Potter, J.A, Owsianka, A, Angus, A.G.N, Jeffery, N, Matthews, D, Keck, Z, Lau, P, Foung, S.K.H, Taylor, G.L, Patel, A.H. | | Deposit date: | 2012-07-25 | | Release date: | 2012-10-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a Hepatitis C Virus Vaccine: the Structural Basis of Hepatitis C Virus Neutralization by AP33, a Broadly Neutralizing Antibody.
J.Virol., 86, 2012
|
|
5BPV
 
 | | Crystal Structure of Zaire ebolavirus VP35 RNA binding domain mutant I278A | | Descriptor: | Polymerase cofactor VP35 | | Authors: | Fadda, V, Cannas, V, Zinzula, L, Distinto, S, Daino, G.L, Bianco, G, Corona, A, Esposito, F, Alcaro, S, Maccioni, E, Tramontano, E, Taylor, G.L. | | Deposit date: | 2015-05-28 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Crystal Structure of Zaire ebolavirus VP35 RNA binding domain mutant I278A
to be published
|
|
4M4U
 
 | |
4XYX
 
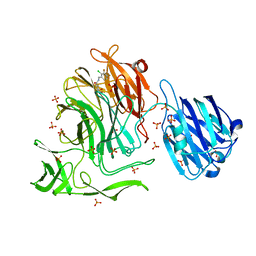 | | NanB plus Optactamide | | Descriptor: | Optactamide, PHOSPHATE ION, Sialidase B | | Authors: | Rogers, G.W, Brear, P, Yang, L, Taylor, G.L, Westwood, N.J. | | Deposit date: | 2015-02-03 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Hunt for Serendipitous Allosteric Sites: Discovery of a novel allosteric inhibitor of the bacterial sialidase NanB
To Be Published
|
|
4M4V
 
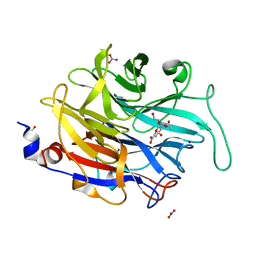 | |
6ER2
 
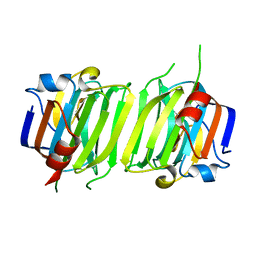 | |
2VVZ
 
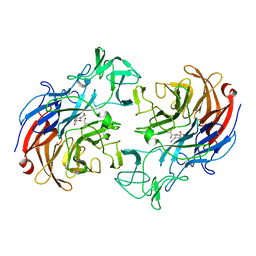 | | Structure of the catalytic domain of Streptococcus pneumoniae sialidase NanA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CHLORIDE ION, SIALIDASE A | | Authors: | Xu, G, Li, X, Andrew, P.W, Taylor, G.L. | | Deposit date: | 2008-06-13 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Catalytic Domain of Streptococcus Pneumoniae Sialidase Nana.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
4M4N
 
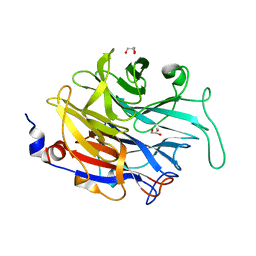 | |
2W20
 
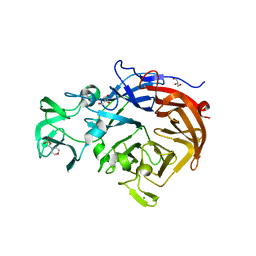 | | Structure of the catalytic domain of the native NanA sialidase from Streptococcus pneumoniae | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Xu, G, Andrew, P.W, Taylor, G.L. | | Deposit date: | 2008-10-21 | | Release date: | 2008-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure of the Catalytic Domain of Streptococcus Pneumoniae Sialidase Nana.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2BER
 
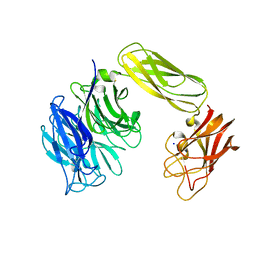 | | Y370G Active Site Mutant of the Sialidase from Micromonospora viridifaciens in complex with beta-Neu5Ac (sialic acid). | | Descriptor: | BACTERIAL SIALIDASE, N-acetyl-beta-neuraminic acid, SODIUM ION | | Authors: | Newstead, S, Watson, J.N, Bennet, A.J, Taylor, G.L. | | Deposit date: | 2004-11-30 | | Release date: | 2005-04-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Mechanism of Action of an Inverting Mutant Sialidase.
Biochemistry, 44, 2005
|
|
1W8N
 
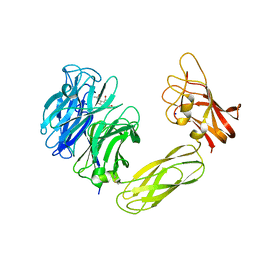 | | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens. | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, BACTERIAL SIALIDASE, SODIUM ION, ... | | Authors: | Newstead, S, Watson, J.N, Dookhun, V, Bennet, A.J, Taylor, G. | | Deposit date: | 2004-09-24 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora Viridifaciens
FEBS Lett., 577, 2004
|
|
2W38
 
 | | Crystal structure of the pseudaminidase from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, SIALIDASE | | Authors: | Xu, G, Ryan, C, Kiefel, M.J, Wilson, J.C, Taylor, G.L. | | Deposit date: | 2008-11-07 | | Release date: | 2008-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies on the Pseudomonas Aeruginosa Sialidase-Like Enzyme Pa2794 Suggest Substrate and Mechanistic Variations.
J.Mol.Biol., 386, 2009
|
|
2VW1
 
 | | Crystal structure of the NanB sialidase from Streptococcus pneumoniae | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, SIALIDASE B | | Authors: | Xu, G, Potter, J.A, Russell, R.J.M, Oggioni, M.R, Andrew, P.W, Taylor, G.L. | | Deposit date: | 2008-06-13 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal Structure of the Nanb Sialidase from Streptococcus Pneumoniae
J.Mol.Biol., 384, 2008
|
|
2VW2
 
 | | Crystal structure of the NanB sialidase from Streptococcus pneumoniae | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, SIALIDASE B | | Authors: | Xu, G, Potter, J.A, Russell, R.J.M, Oggioni, M.R, Andrew, P.W, Taylor, G.L. | | Deposit date: | 2008-06-13 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Nanb Sialidase from Streptococcus Pneumoniae
J.Mol.Biol., 384, 2008
|
|
