3IGL
 
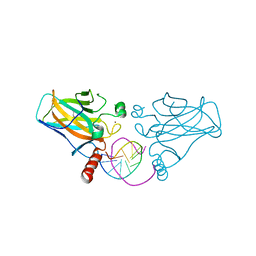 | | Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs (p53-DNA complex 1) | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DNA (5'-D(*CP*GP*GP*GP*CP*AP*TP*GP*CP*CP*CP*G)-3'), ... | | Authors: | Kitayner, M, Suad, O, Rozenberg, H, Shakked, Z. | | Deposit date: | 2009-07-28 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs
Nat.Struct.Mol.Biol., 17, 2010
|
|
2ADY
 
 | | Structural Basis of DNA Recognition by p53 Tetramers (complex IV) | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*AP*TP*GP*TP*CP*CP*G)-3', Cellular tumor antigen p53, ZINC ION | | Authors: | Kitayner, M, Rozenberg, H, Kessler, N, Rabinovich, D, Shakked, Z. | | Deposit date: | 2005-07-21 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of DNA Recognition by p53 Tetramers
Mol.Cell, 22, 2006
|
|
2ATA
 
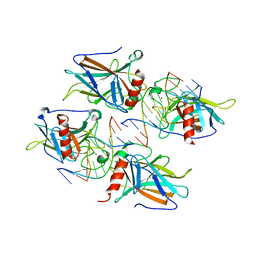 | | Structural Basis of DNA Recognition by p53 Tetramers (complex II) | | Descriptor: | 5'-D(*AP*AP*GP*GP*CP*AP*TP*GP*CP*CP*TP*T)-3', Cellular tumor antigen p53, ZINC ION | | Authors: | Kitayner, M, Rozenberg, H, Kessler, N, Rabinovich, D, Shakked, Z. | | Deposit date: | 2005-08-24 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of DNA Recognition by p53 Tetramers
Mol.Cell, 22, 2006
|
|
2AC0
 
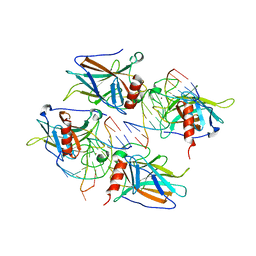 | | Structural Basis of DNA Recognition by p53 Tetramers (complex I) | | Descriptor: | 5'-D(*CP*GP*GP*GP*CP*AP*TP*GP*CP*CP*CP*G)-3', Cellular tumor antigen p53, ZINC ION | | Authors: | Kitayner, M, Rozenberg, H, Kessler, N, Rabinovich, D, Shakked, Z. | | Deposit date: | 2005-07-18 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of DNA Recognition by p53 Tetramers
Mol.Cell, 22, 2006
|
|
2AHI
 
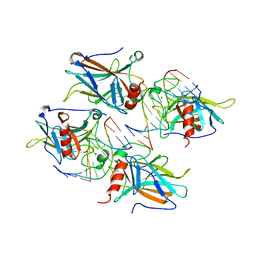 | | Structural Basis of DNA Recognition by p53 Tetramers (complex III) | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*AP*TP*GP*TP*CP*CP*G)-3', Cellular tumor antigen p53, ZINC ION | | Authors: | Kitayner, M, Rozenberg, H, Kessler, N, Rabinovich, D, Shakked, Z. | | Deposit date: | 2005-07-28 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of DNA Recognition by p53 Tetramers
Mol.Cell, 22, 2006
|
|
1XJY
 
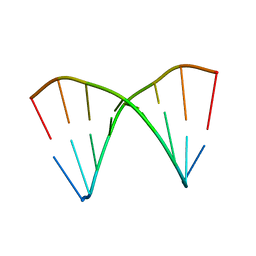 | | The crystal structures of the DNA binding sites of the RUNX1 transcription factor | | Descriptor: | 5'-D(*TP*CP*TP*GP*CP*GP*GP*TP*C)-3', 5'-D(*TP*GP*AP*CP*CP*GP*CP*AP*G)-3' | | Authors: | Kitayner, M, Rozenberg, H, Rabinovich, D, Shakked, Z. | | Deposit date: | 2004-09-26 | | Release date: | 2005-03-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the DNA-binding site of Runt-domain transcription regulators.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1XJX
 
 | | The crystal structures of the DNA binding sites of the RUNX1 transcription factor | | Descriptor: | 5'-D(*TP*CP*TP*GP*CP*GP*GP*TP*C)-3', 5'-D(*TP*GP*AP*CP*CP*GP*CP*AP*G)-3' | | Authors: | Kitayner, M, Rozenberg, H, Rabinovich, D, Shakked, Z. | | Deposit date: | 2004-09-26 | | Release date: | 2005-03-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the DNA-binding site of Runt-domain transcription regulators.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5NAK
 
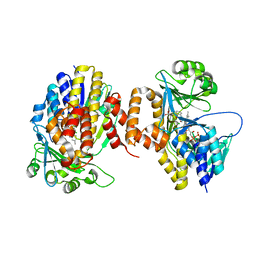 | | Pseudomonas fluorescens kynurenine 3-monooxygenase (KMO) in complex with the enzyme substrate L-kynurenine | | Descriptor: | (2S)-2-amino-4-(2-aminophenyl)-4-oxobutanoic acid, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Taylor, M, Mowat, C.G, Rowland, P. | | Deposit date: | 2017-02-28 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic basis of differentiated inhibitors of the acute pancreatitis target kynurenine-3-monooxygenase.
Nat Commun, 8, 2017
|
|
3WVO
 
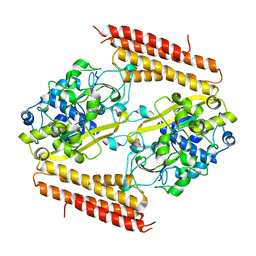 | | Crystal structure of Thermobifida fusca Cse1 | | Descriptor: | CRISPR-associated protein, Cse1 family | | Authors: | Yuan, Y.A, Tay, M. | | Deposit date: | 2014-06-02 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal structure of Thermobifida fusca Cse1 reveals target DNA binding site.
Protein Sci., 24, 2015
|
|
1W0P
 
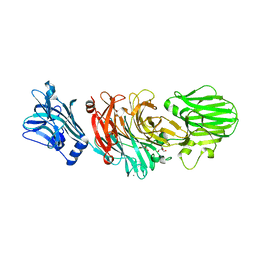 | | Vibrio cholerae sialidase with alpha-2,6-sialyllactose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Moustafa, I, Connaris, H, Taylor, M, Zaitsev, V, Wilson, J.C, Kiefel, M.J, von-Itzstein, M, Taylor, G. | | Deposit date: | 2004-06-09 | | Release date: | 2004-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Sialic Acid Recognition by Vibrio Cholerae Neuraminidase.
J.Biol.Chem., 279, 2004
|
|
1W0O
 
 | | Vibrio cholerae sialidase | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Moustafa, I, Connaris, H, Taylor, M, Zaitsev, V, Wilson, J.C, Kiefel, M.J, von-Itzstein, M, Taylor, G. | | Deposit date: | 2004-06-09 | | Release date: | 2004-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sialic Acid Recognition by Vibrio Cholerae Neuraminidase
J.Biol.Chem., 279, 2004
|
|
1NY3
 
 | | Crystal structure of ADP bound to MAP KAP kinase 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAP kinase-activated protein kinase 2 | | Authors: | Underwood, K.W, Parris, K.D, Federico, E, Mosyak, L, Shane, T, Taylor, M, Svenson, K, Liu, Y, Hsiao, C.L, Wolfrom, S, Maguire, M, Malakian, K, Telliez, J.B, Lin, L.L, Kriz, R.W, Seehra, J, Somers, W.S, Stahl, M.L. | | Deposit date: | 2003-02-11 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Catalytically active MAP KAP kinase 2 structures in complex with staurosporine and ADP reveal differences with the autoinhibited enzyme
Structure, 11, 2003
|
|
1S1E
 
 | | Crystal Structure of Kv Channel-interacting protein 1 (KChIP-1) | | Descriptor: | CALCIUM ION, Kv channel interacting protein 1 | | Authors: | Scannevin, R.H, Wang, K.-W, Jow, F, Megules, J, Kopsco, D.C, Edris, W, Carroll, K.C, Lu, Q, Xu, W.-X, Xu, Z.-B, Katz, A.H, Olland, S, Lin, L, Taylor, M, Stahl, M, Malakian, K, Somers, W, Mosyak, L, Bowlby, M.R, Chanda, P, Rhodes, K.J. | | Deposit date: | 2004-01-06 | | Release date: | 2005-01-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two N-terminal domains of Kv4 K(+) channels regulate binding to and modulation by KChIP1.
Neuron, 41, 2004
|
|
1S1G
 
 | | Crystal Structure of Kv4.3 T1 Domain | | Descriptor: | Potassium voltage-gated channel subfamily D member 3, ZINC ION | | Authors: | Scannevin, R.H, Wang, K.W, Jow, F, Megules, J, Kopsco, D.C, Edris, W, Carroll, K.C, Lu, Q, Xu, W.X, Xu, Z.B, Katz, A.H, Olland, S, Lin, L, Taylor, M, Stahl, M, Malakian, K, Somers, W, Mosyak, L, Bowlby, M.R, Chanda, P, Rhodes, K.J. | | Deposit date: | 2004-01-06 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Two N-terminal domains of Kv4 K(+) channels regulate binding to and modulation by KChIP1.
Neuron, 41, 2004
|
|
1NXK
 
 | | Crystal structure of staurosporine bound to MAP KAP kinase 2 | | Descriptor: | MAP kinase-activated protein kinase 2, STAUROSPORINE, SULFATE ION | | Authors: | Underwood, K.W, Parris, K.D, Federico, E, Mosyak, L, Czerwinski, R.M, Shane, T, Taylor, M, Svenson, K, Liu, Y, Hsiao, C.L, Wolfrom, S, Malakian, K, Telliez, J.B, Lin, L.L, Kriz, R.W, Seehra, J, Somers, W.S, Stahl, M.L. | | Deposit date: | 2003-02-10 | | Release date: | 2003-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Catalytically active MAP KAP kinase 2 structures in complex with staurosporine and ADP reveal differences with the autoinhibited enzyme
Structure, 11, 2003
|
|
3AUK
 
 | | Crystal structure of a lipase from Geobacillus sp. SBS-4S | | Descriptor: | CALCIUM ION, CHLORIDE ION, Lipase, ... | | Authors: | Angkawidjaja, C, Tayyab, M, Rashid, N, Kanaya, S. | | Deposit date: | 2011-02-08 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of a lipase from Geobacillus sp. SBS-4S
TO BE PUBLISHED
|
|
5AUK
 
 | | Crystal structure of the Ga-substituted Ferredoxin | | Descriptor: | BENZAMIDINE, Ferredoxin-1, SULFATE ION, ... | | Authors: | Kurisu, G, Muraki, N, Taya, M, Hase, T. | | Deposit date: | 2015-04-24 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | X-ray Structure and Nuclear Magnetic Resonance Analysis of the Interaction Sites of the Ga-Substituted Cyanobacterial Ferredoxin
Biochemistry, 54, 2015
|
|
