6C7S
 
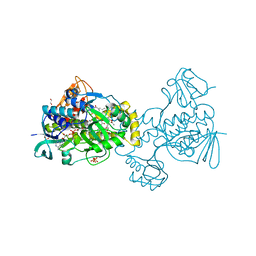 | | Structure of Rifampicin Monooxygenase with Product Bound | | Descriptor: | (1E,3S,4R,5S,6R,7R,8R,9S,10S,11E,13E)-15-amino-1-{[(2S)-5,7-dihydroxy-2,4-dimethyl-8-{(E)-[(4-methylpiperazin-1-yl)imino]methyl}-1,6,9-trioxo-1,2,6,9-tetrahydronaphtho[2,1-b]furan-2-yl]oxy}-7,9-dihydroxy-3-methoxy-4,6,8,10,14-pentamethyl-15-oxopentadeca-1,11,13-trien-5-yl acetate, 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Liu, L.-K, Tanner, J.J. | | Deposit date: | 2018-01-23 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Evidence for Rifampicin Monooxygenase Inactivating Rifampicin by Cleaving Its Ansa-Bridge.
Biochemistry, 57, 2018
|
|
6D96
 
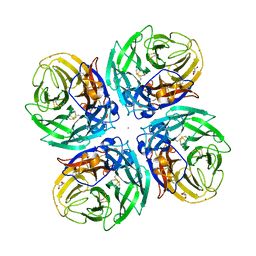 | |
4U8I
 
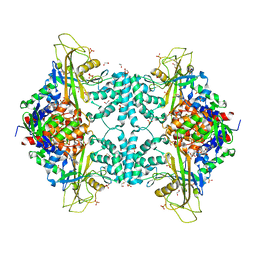 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant F66A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8P
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Y317A complexed with UDP | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8O
 
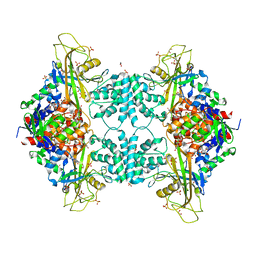 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant N207A complexed with UDP | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8L
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant N207A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8N
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant F66A complexed with UDP | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8M
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Y317A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8K
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Q107A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8J
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Y104A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
8DKG
 
 | | Structure of PYCR1 Thr171Met variant complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Isoform 3 of Pyrroline-5-carboxylate reductase 1, mitochondrial, ... | | Authors: | Meeks, K.R, Tanner, J.J. | | Deposit date: | 2022-07-05 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Functional Impact of a Cancer-Related Variant in Human Delta 1 -Pyrroline-5-Carboxylate Reductase 1.
Acs Omega, 8, 2023
|
|
1U8F
 
 | |
4X0U
 
 | | Structure ALDH7A1 inactivated by 4-diethylaminobenzaldehyde | | Descriptor: | 4-(diethylamino)benzaldehyde, Alpha-aminoadipic semialdehyde dehydrogenase, MAGNESIUM ION | | Authors: | Luo, M, Tanner, J.J. | | Deposit date: | 2014-11-23 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Diethylaminobenzaldehyde Is a Covalent, Irreversible Inactivator of ALDH7A1.
Acs Chem.Biol., 10, 2015
|
|
4X0T
 
 | |
1XF3
 
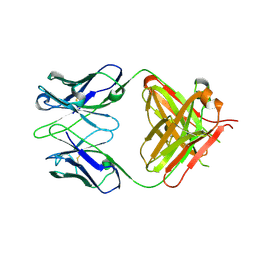 | | Structure of ligand-free Fab DNA-1 in space group P65 | | Descriptor: | Fab Light chain, Fab heavy chain | | Authors: | Schuermann, J.P, Prewitt, S.P, Deutscher, S.L, Tanner, J.J. | | Deposit date: | 2004-09-13 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence for Structural Plasticity of Heavy Chain Complementarity-determining Region 3 in Antibody-ssDNA Recognition
J.Mol.Biol., 347, 2005
|
|
1XF4
 
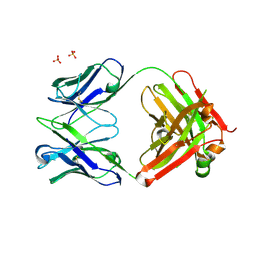 | | Structure of ligand-free Fab DNA-1 in space group P321 solved from crystals with perfect hemihedral twinning | | Descriptor: | Fab heavy chain, Fab light chain, SULFATE ION | | Authors: | Schuermann, J.P, Prewitt, S.P, Deutscher, S.L, Tanner, J.J. | | Deposit date: | 2004-09-13 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evidence for Structural Plasticity of Heavy Chain Complementarity-determining Region 3 in Antibody-ssDNA Recognition
J.Mol.Biol., 347, 2005
|
|
1XF2
 
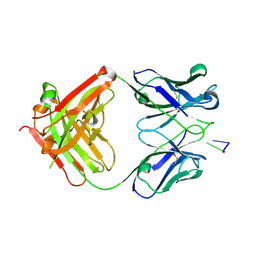 | | Structure of Fab DNA-1 complexed with dT3 | | Descriptor: | 5'-D(*TP*TP*T)-3', SULFATE ION, antibody heavy chain Fab, ... | | Authors: | Schuermann, J.P, Prewitt, S.P, Deutscher, S.L, Tanner, J.J. | | Deposit date: | 2004-09-13 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence for Structural Plasticity of Heavy Chain Complementarity-determining Region 3 in Antibody-ssDNA Recognition
J.Mol.Biol., 347, 2005
|
|
3FS7
 
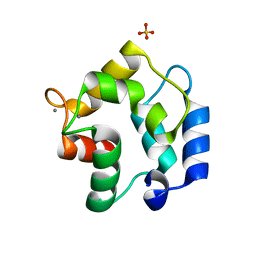 | | Crystal structure of Gallus gallus beta-parvalbumin (avian thymic hormone) | | Descriptor: | CALCIUM ION, GLYCEROL, Parvalbumin, ... | | Authors: | Schuermann, J.P, Tanner, J.J, Henzl, M.T. | | Deposit date: | 2009-01-09 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9539 Å) | | Cite: | Structure of avian thymic hormone, a high-affinity avian beta-parvalbumin, in the Ca2+-free and Ca2+-bound states.
J.Mol.Biol., 397, 2010
|
|
3IT3
 
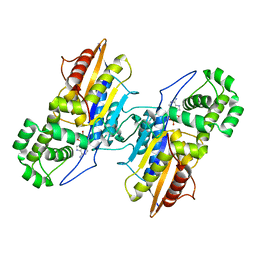 | | Crystal Structure Francisella tularensis histidine acid phosphatase D261A mutant complexed with substrate 3'-AMP | | Descriptor: | Acid phosphatase, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl] dihydrogen phosphate | | Authors: | Singh, H, Felts, R.L, Reilly, T.J, Tanner, J.J. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of the histidine acid phosphatase from Francisella tularensis provide insight into substrate recognition.
J.Mol.Biol., 394, 2009
|
|
3IT1
 
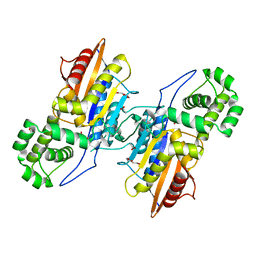 | | Crystal Structure Francisella tularensis histidine acid phosphatase complexed with L(+)-tartrate | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, ACETATE ION, Acid phosphatase, ... | | Authors: | Singh, H, Felts, R.L, Reilly, T.J, Tanner, J.J. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.691 Å) | | Cite: | Crystal Structures of the histidine acid phosphatase from Francisella tularensis provide insight into substrate recognition.
J.Mol.Biol., 394, 2009
|
|
3IT0
 
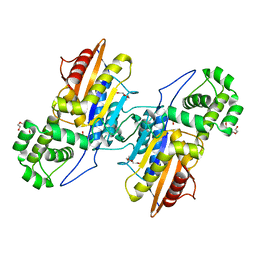 | | Crystal Structure Francisella tularensis histidine acid phosphatase complexed with phosphate | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Acid phosphatase, PENTAETHYLENE GLYCOL, ... | | Authors: | Singh, H, Felts, R.L, Reilly, T.J, Tanner, J.J. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Crystal Structures of the histidine acid phosphatase from Francisella tularensis provide insight into substrate recognition.
J.Mol.Biol., 394, 2009
|
|
3IT2
 
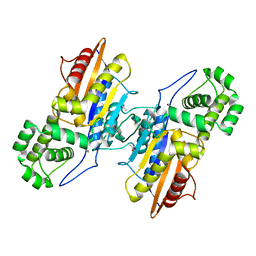 | | Crystal structure of ligand-free Francisella tularensis histidine acid phosphatase | | Descriptor: | ACETATE ION, Acid phosphatase | | Authors: | Singh, H, Felts, R.L, Reilly, T.J, Tanner, J.J. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.838 Å) | | Cite: | Crystal Structures of the histidine acid phosphatase from Francisella tularensis provide insight into substrate recognition.
J.Mol.Biol., 394, 2009
|
|
6UFP
 
 | | Structure of proline utilization A with the FAD covalently modified by L-thiazolidine-2-carboxylate and three cysteines (Cys46, Cys470, Cys638) modified to S,S-(2-HYDROXYETHYL)THIOCYSTEINE | | Descriptor: | (2S)-1,3-thiazolidine-2-carboxylic acid, Bifunctional protein PutA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Campbell, A.C, Tanner, J.J. | | Deposit date: | 2019-09-24 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | Covalent Modification of the Flavin in Proline Dehydrogenase by Thiazolidine-2-Carboxylate.
Acs Chem.Biol., 15, 2020
|
|
6UXK
 
 | |
6UXI
 
 | | Structure of serine hydroxymethyltransferase 8 from Glycine max cultivar Essex complexed with PLP-Glycine | | Descriptor: | 1,2-ETHANEDIOL, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], Serine hydroxymethyltransferase | | Authors: | Korasick, D.A, Tanner, J.J, Beamer, L.J. | | Deposit date: | 2019-11-07 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Impaired folate binding of serine hydroxymethyltransferase 8 from soybean underlies resistance to the soybean cyst nematode.
J.Biol.Chem., 295, 2020
|
|
