6JNJ
 
 | |
1ITU
 
 | | HUMAN RENAL DIPEPTIDASE COMPLEXED WITH CILASTATIN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CILASTATIN, RENAL DIPEPTIDASE, ... | | 著者 | Nitanai, Y, Satow, Y, Adachi, H, Tsujimoto, M. | | 登録日 | 2002-02-03 | | 公開日 | 2002-08-28 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of Human Renal Dipeptidase Involved in beta-Lactam Hydrolysis
J.Mol.Biol., 321, 2002
|
|
1C4L
 
 | | SOLUTION STRUCTURE OF AN RNA DUPLEX INCLUDING A C-U BASE-PAIR | | 分子名称: | RNA (5'-R(*CP*CP*UP*GP*CP*GP*UP*CP*G)-3'), RNA (5'-R(*CP*GP*AP*CP*UP*CP*AP*GP*G)-3') | | 著者 | Tanaka, Y, Kojima, C, Yamazaki, T, Kodama, T.S, Yasuno, K, Miyashita, S, Ono, A.M, Ono, A.S, Kainosho, M, Kyogoku, Y. | | 登録日 | 1999-08-30 | | 公開日 | 2000-08-09 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of an RNA duplex including a C-U base pair.
Biochemistry, 39, 2000
|
|
1ITQ
 
 | | HUMAN RENAL DIPEPTIDASE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RENAL DIPEPTIDASE, ZINC ION | | 著者 | Nitanai, Y, Satow, Y, Adachi, H, Tsujimoto, M. | | 登録日 | 2002-02-02 | | 公開日 | 2002-08-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structure of Human Renal Dipeptidase Involved in beta-Lactam Hydrolysis
J.Mol.Biol., 321, 2002
|
|
2Z6F
 
 | |
6L06
 
 | |
5WED
 
 | |
6L07
 
 | |
3F95
 
 | | Crystal Structure of Extra C-terminal Domain (X) of Exo-1,3/1,4-beta-glucanase (ExoP) from Pseudoalteromonas sp. BB1 | | 分子名称: | Beta-glucosidase, CHLORIDE ION | | 著者 | Nakatani, Y, Cutfield, S.M, Cutfield, J.F. | | 登録日 | 2008-11-13 | | 公開日 | 2009-11-17 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure and activity of exo-1,3/1,4-beta-glucanase from marine bacterium Pseudoalteromonas sp. BB1 showing a novel C-terminal domain
Febs J., 2011
|
|
6JNK
 
 | |
3Q35
 
 | | Structure of the Rtt109-AcCoA/Vps75 complex and implications for chaperone-mediated histone acetylation | | 分子名称: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, Histone acetyltransferase, ... | | 著者 | Tang, Y, Yuan, H, Meeth, K, Marmorstein, R. | | 登録日 | 2010-12-21 | | 公開日 | 2011-02-02 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structure of the Rtt109-AcCoA/Vps75 Complex and Implications for Chaperone-Mediated Histone Acetylation.
Structure, 19, 2011
|
|
6K9Y
 
 | | Crystal structure of human VAT-1 | | 分子名称: | NITRATE ION, Synaptic vesicle membrane protein VAT-1 homolog | | 著者 | Watanabe, Y, Endo, T. | | 登録日 | 2019-06-19 | | 公開日 | 2020-02-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis for interorganelle phospholipid transport mediated by VAT-1.
J.Biol.Chem., 295, 2020
|
|
3Q33
 
 | | Structure of the Rtt109-AcCoA/Vps75 Complex and Implications for Chaperone-Mediated Histone Acetylation | | 分子名称: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, HISTONE H3, ... | | 著者 | Tang, Y, Yuan, H, Meeth, K, Marmorstein, R. | | 登録日 | 2010-12-21 | | 公開日 | 2011-02-02 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of the Rtt109-AcCoA/Vps75 Complex and Implications for Chaperone-Mediated Histone Acetylation.
Structure, 19, 2011
|
|
5HEE
 
 | | Crystal structure of the TK2203 protein | | 分子名称: | GLYCEROL, Putative uncharacterized protein, TK2203 protein, ... | | 著者 | Nishitani, Y, Miki, K. | | 登録日 | 2016-01-06 | | 公開日 | 2016-06-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Crystal structure of the TK2203 protein from Thermococcus kodakarensis, a putative extradiol dioxygenase
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6IUR
 
 | | A phosphatase complex STRN3-PP2Aa | | 分子名称: | PP2A scaffolding subunit, PROPANE, Striatin-3 | | 著者 | Tang, Y, Zhou, Z.C. | | 登録日 | 2018-11-30 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.33 Å) | | 主引用文献 | Selective Inhibition of STRN3-Containing PP2A Phosphatase Restores Hippo Tumor-Suppressor Activity in Gastric Cancer.
Cancer Cell, 38, 2020
|
|
4YTV
 
 | | Crystal structure of Mdm35 | | 分子名称: | COBALT (II) ION, GLYCEROL, Mitochondrial distribution and morphology protein 35 | | 著者 | Watanabe, Y, Tamura, Y, Kawano, S, Endo, T. | | 登録日 | 2015-03-18 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria.
Nat Commun, 6, 2015
|
|
2HG4
 
 | | Structure of the ketosynthase-acyltransferase didomain of module 5 from DEBS. | | 分子名称: | 6-Deoxyerythronolide B Synthase, ACETATE ION, CHLORIDE ION, ... | | 著者 | Tang, Y, Kim, C.Y, Mathews, I.I, Cane, D.E, Khosla, C. | | 登録日 | 2006-06-26 | | 公開日 | 2006-07-11 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | The 2.7-A crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4YTW
 
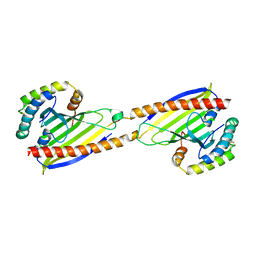 | | Crystal structure of Ups1-Mdm35 complex | | 分子名称: | Mitochondrial distribution and morphology protein 35, Protein UPS1, mitochondrial | | 著者 | Watanabe, Y, Tamura, Y, Kawano, S, Endo, T. | | 登録日 | 2015-03-18 | | 公開日 | 2015-08-12 | | 最終更新日 | 2020-02-05 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria.
Nat Commun, 6, 2015
|
|
5B13
 
 | |
4YTX
 
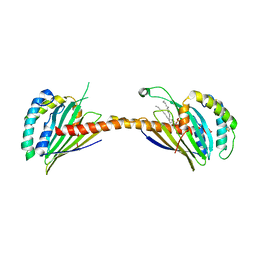 | | Crystal structure of Ups1-Mdm35 complex with PA | | 分子名称: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, Mitochondrial distribution and morphology protein 35, Protein UPS1, ... | | 著者 | Watanabe, Y, Tamura, Y, Kawano, S, Endo, T. | | 登録日 | 2015-03-18 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria.
Nat Commun, 6, 2015
|
|
5AZF
 
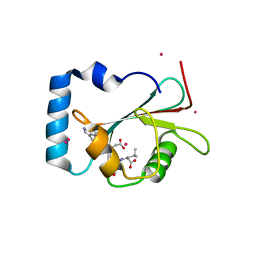 | | Crystal structure of LGG-1 complexed with a WEEL peptide | | 分子名称: | CADMIUM ION, Protein lgg-1, SULFATE ION, ... | | 著者 | Watanabe, Y, Noda, N.N. | | 登録日 | 2015-10-05 | | 公開日 | 2015-12-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy.
Mol.Cell, 60, 2015
|
|
5AZH
 
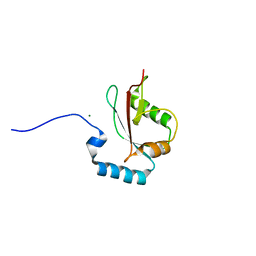 | | Crystal structure of LGG-2 fused with an EEEWEEL peptide | | 分子名称: | EEEWEEL peptide,Protein lgg-2, MAGNESIUM ION | | 著者 | Watanabe, Y, Fujioka, Y, Noda, N.N. | | 登録日 | 2015-10-05 | | 公開日 | 2015-12-30 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy.
Mol.Cell, 60, 2015
|
|
3RRX
 
 | | Crystal Structure of Q683A mutant of Exo-1,3/1,4-beta-glucanase (ExoP) from Pseudoalteromonas sp. BB1 | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Exo-1,3/1,4-beta-glucanase, ... | | 著者 | Nakatani, Y, Cutfield, S.M, Cutfield, J.F. | | 登録日 | 2011-05-01 | | 公開日 | 2011-12-21 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and activity of exo-1,3/1,4-beta-glucanase from marine bacterium Pseudoalteromonas sp. BB1 showing a novel C-terminal domain
Febs J., 279, 2012
|
|
2D3E
 
 | |
2JRM
 
 | | Solution NMR structure of ribosome modulation factor VP1593 from Vibrio parahaemolyticus. Northeast Structural Genomics target VpR55 | | 分子名称: | Ribosome modulation factor | | 著者 | Tang, Y, Rossi, P, Swapna, G, Wang, H, Jiang, M, Cunningham, K, Owens, L, Ma, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-06-27 | | 公開日 | 2007-07-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure of Ribosome Modulation Factor VP1593 from Vibrio parahaemolyticus.
To be Published
|
|
