7CGR
 
 | |
7DO6
 
 | |
7FGP
 
 | |
7CGQ
 
 | |
7EQF
 
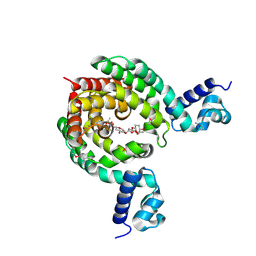 | | Crystal Structure of a Transcription Factor in complex with Ligand | | Descriptor: | (6~{R})-3-methyl-8-[(2~{S},4~{R},5~{S},6~{R})-6-methyl-5-[(2~{S},4~{R},5~{R},6~{R})-6-methyl-4-[(2~{S},5~{S},6~{S})-6-methyl-5-[(2~{S},4~{R},5~{S},6~{R})-6-methyl-5-[(2~{S},4~{S},5~{S},6~{R})-6-methyl-4-[(2~{S},5~{S},6~{S})-6-methyl-5-oxidanyl-oxan-2-yl]oxy-5-oxidanyl-oxan-2-yl]oxy-4-oxidanyl-oxan-2-yl]oxy-oxan-2-yl]oxy-5-oxidanyl-oxan-2-yl]oxy-4-oxidanyl-oxan-2-yl]oxy-1,6,11-tris(oxidanyl)-5,6-dihydrobenzo[a]anthracene-7,12-dione, TetR/AcrR family transcriptional regulator | | Authors: | Uehara, S, Tsugita, A, Matsui, T, Yokoyama, T, Ostash, I, Ostash, B, Tanaka, Y. | | Deposit date: | 2021-05-01 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | The carbohydrate tail of landomycin A is responsible for its interaction with the repressor protein LanK.
Febs J., 289, 2022
|
|
7DO5
 
 | |
7DO7
 
 | |
7CNQ
 
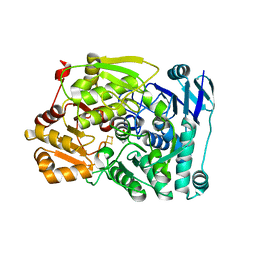 | | Crystal structure of Agrobacterium tumefaciens aconitase X (holo-form) | | Descriptor: | (2~{S},3~{R})-3-oxidanylpyrrolidine-2-carboxylic acid, FE2/S2 (INORGANIC) CLUSTER, cis-3-hydroxy-L-proline dehydratase | | Authors: | Murase, Y, Watanabe, Y, Watanabe, S. | | Deposit date: | 2020-08-03 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of aconitase X enzymes from bacteria and archaea provide insights into the molecular evolution of the aconitase superfamily.
Commun Biol, 4, 2021
|
|
7CNP
 
 | |
7CNS
 
 | | Crystal structure of Thermococcus kodakaraensis aconitase X (holo-form) | | Descriptor: | (3R)-3-HYDROXY-3-METHYL-5-(PHOSPHONOOXY)PENTANOIC ACID, DUF521 domain-containing protein, FE3-S4 CLUSTER, ... | | Authors: | Murase, Y, Watanabe, Y, Watanabe, S. | | Deposit date: | 2020-08-03 | | Release date: | 2021-06-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal structures of aconitase X enzymes from bacteria and archaea provide insights into the molecular evolution of the aconitase superfamily.
Commun Biol, 4, 2021
|
|
7CNR
 
 | |
7D2R
 
 | | Crystal structure of Agrobacterium tumefaciens aconitase X mutant - S449C/C510V | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, SODIUM ION, ... | | Authors: | Murase, Y, Watanabe, Y, Watanabe, S. | | Deposit date: | 2020-09-17 | | Release date: | 2021-06-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Crystal structures of aconitase X enzymes from bacteria and archaea provide insights into the molecular evolution of the aconitase superfamily.
Commun Biol, 4, 2021
|
|
6ZEC
 
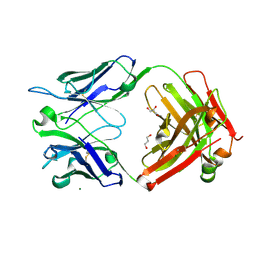 | | Crystal Structure of the Fab Fragment of a Glycosylated Lymphoma Antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab fragment heavy chain, Fab fragment light chain, ... | | Authors: | Allen, J.D, Watanabe, Y, Crispin, M, Bowden, T.A. | | Deposit date: | 2020-06-16 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insertion of atypical glycans into the tumor antigen-binding site identifies DLBCLs with distinct origin and behavior.
Blood, 138, 2021
|
|
6HJ5
 
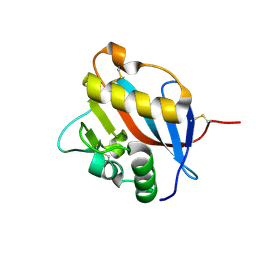 | | Crystal structure of Whitewater Arroyo virus GP1 glycoprotein at pH 5.6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pre-glycoprotein polyprotein GP complex | | Authors: | Pryce, R, Ng, W.M, Zeltina, A, Watanabe, Y, El Omari, K, Wagner, A, Bowden, T.A. | | Deposit date: | 2018-08-31 | | Release date: | 2018-10-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure-Based Classification Defines the Discrete Conformational Classes Adopted by the Arenaviral GP1.
J. Virol., 93, 2019
|
|
6HJC
 
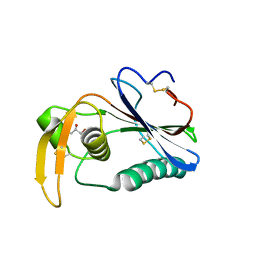 | | Crystal structure of Loei River virus GP1 glycoprotein at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Pre-glycoprotein polyprotein GP complex | | Authors: | Pryce, R, Ng, W.M, Zeltina, A, Watanabe, Y, El Omari, K, Wagner, A, Bowden, T.A. | | Deposit date: | 2018-09-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure-Based Classification Defines the Discrete Conformational Classes Adopted by the Arenaviral GP1.
J. Virol., 93, 2019
|
|
6HJ6
 
 | | Crystal structure of Loei River virus GP1 glycoprotein at pH 5.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Pre-glycoprotein polyprotein GP complex | | Authors: | Pryce, R, Ng, W.M, Zeltina, A, Watanabe, Y, El Omari, K, Wagner, A, Bowden, T.A. | | Deposit date: | 2018-08-31 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-Based Classification Defines the Discrete Conformational Classes Adopted by the Arenaviral GP1.
J. Virol., 93, 2019
|
|
7EQE
 
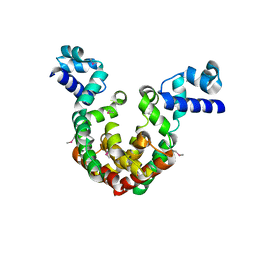 | | Crystal Structure of a transcription factor | | Descriptor: | TetR/AcrR family transcriptional regulator | | Authors: | Uehara, S, Tsugita, A, Matsui, T, Yokoyama, T, Ostash, I, Ostash, B, Tanaka, Y. | | Deposit date: | 2021-05-01 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | The carbohydrate tail of landomycin A is responsible for its interaction with the repressor protein LanK.
Febs J., 289, 2022
|
|
6AJA
 
 | | Crystal structure of Trypanosoma brucei glycosomal isocitrate dehydrogenase in complex with NADPH, alpha-ketoglutarate and ca2+ | | Descriptor: | 2-OXOGLUTARIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, X, Inaoka, D.K, Shiba, T, Balogun, E.O, Ziebart, N, Allman, S, Watanabe, Y, Nozaki, T, Boshart, M, Bringaud, F, Harada, S, Kita, K. | | Deposit date: | 2018-08-27 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Biochemical characterization of a novel Trypanosoma brucei glycosomal isocitrate dehydrogenase with dual coenzyme specificity (NADP+/NAD+)
To Be Published
|
|
6AJB
 
 | | Crystal structure of Trypanosoma brucei glycosomal isocitrate dehydrogenase in complex with NADH, alpha-ketoglutarate and ca2+ | | Descriptor: | 2-OXOGLUTARIC ACID, CALCIUM ION, Isocitrate dehydrogenase [NADP], ... | | Authors: | Wang, X, Inaoka, D.K, Shiba, T, Balogun, E.O, Ziebart, N, Allman, S, Watanabe, Y, Nozaki, T, Boshart, M, Bringaud, F, Harada, S, Kita, K. | | Deposit date: | 2018-08-27 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Biochemical characterization of a novel Trypanosoma brucei glycosomal isocitrate dehydrogenase with dual coenzyme specificity (NADP+/NAD+)
To Be Published
|
|
6AJ6
 
 | | Crystal structure of Trypanosoma brucei glycosomal isocitrate dehydrogenase in complex with NADP+ | | Descriptor: | Isocitrate dehydrogenase [NADP], NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, X, Inaoka, D.K, Shiba, T, Balogun, E.O, Ziebart, N, Allman, S, Watanabe, Y, Nozaki, T, Boshart, M, Bringaud, F, Harada, S, Kita, K. | | Deposit date: | 2018-08-27 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical characterization of a novel Trypanosoma brucei glycosomal isocitrate dehydrogenase with dual coenzyme specificity (NADP+/NAD+)
To Be Published
|
|
6AJ8
 
 | | Crystal structure of Trypanosoma brucei glycosomal isocitrate dehydrogenase in complex with NADP+, alpha-ketoglutarate and ca2+ | | Descriptor: | 2-OXOGLUTARIC ACID, CALCIUM ION, Isocitrate dehydrogenase [NADP], ... | | Authors: | Wang, X, Inaoka, D.K, Shiba, T, Balogun, E.O, Ziebart, N, Allman, S, Watanabe, Y, Nozaki, T, Boshart, M, Bringaud, F, Harada, S, Kita, K. | | Deposit date: | 2018-08-27 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical characterization of a novel Trypanosoma brucei glycosomal isocitrate dehydrogenase with dual coenzyme specificity (NADP+/NAD+)
To Be Published
|
|
2D6Y
 
 | | Crystal Structure of transcriptional factor SCO4008 from Streptomyces coelicolor A3(2) | | Descriptor: | L(+)-TARTARIC ACID, putative tetR family regulatory protein | | Authors: | Hayashi, T, Tanaka, Y, Sakai, N, Yao, M, Tamura, T, Tanaka, I. | | Deposit date: | 2005-11-15 | | Release date: | 2006-10-31 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SCO4008, a Putative TetR Transcriptional Repressor from Streptomyces coelicolor A3(2), Regulates Transcription of sco4007 by Multidrug Recognition.
J.Mol.Biol., 425, 2013
|
|
1IU3
 
 | | CRYSTAL STRUCTURE OF THE E.COLI SEQA PROTEIN COMPLEXED WITH HEMIMETHYLATED DNA | | Descriptor: | 5'-D(*AP*AP*GP*GP*AP*TP*CP*CP*AP*A)-3', 5'-D(*TP*TP*GP*GP*AP*TP*CP*CP*TP*T)-3', SeqA protein | | Authors: | Fujikawa, N, Kurumizaka, H, Nureki, O, Tanaka, Y, Yamazoe, M, Hiraga, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-02-26 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biochemical analyses of hemimethylated DNA binding by the SeqA protein.
Nucleic Acids Res., 32, 2004
|
|
1WSC
 
 | | Crystal structure of ST0229, function unknown protein from Sulfolobus tokodaii | | Descriptor: | Hypothetical protein ST0229 | | Authors: | Murayama, T, Tanaka, Y, Sasaki, T, Yasutake, Y, Yao, M, Tsumoto, K, Tanaka, I, Kumagai, I. | | Deposit date: | 2004-11-05 | | Release date: | 2005-11-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of ST0229, function unknown protein from Sulfolobus tokodaii
To be Published
|
|
6J3N
 
 | | RORgammat LBD complexed with Ursonic Acid and SRC2.2 | | Descriptor: | (5beta)-3-oxours-12-en-28-oic acid, LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN, Nuclear receptor ROR-gamma | | Authors: | Liu, Z.H, Huang, J, Tang, Y. | | Deposit date: | 2019-01-05 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of human RORgammat LBD with SCR2.2 at 1.99 Angstroms resolution
To Be Published
|
|
