4RF2
 
 | |
4RF4
 
 | |
4YTV
 
 | | Crystal structure of Mdm35 | | Descriptor: | COBALT (II) ION, GLYCEROL, Mitochondrial distribution and morphology protein 35 | | Authors: | Watanabe, Y, Tamura, Y, Kawano, S, Endo, T. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria.
Nat Commun, 6, 2015
|
|
4YTW
 
 | | Crystal structure of Ups1-Mdm35 complex | | Descriptor: | Mitochondrial distribution and morphology protein 35, Protein UPS1, mitochondrial | | Authors: | Watanabe, Y, Tamura, Y, Kawano, S, Endo, T. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-12 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria.
Nat Commun, 6, 2015
|
|
3Q35
 
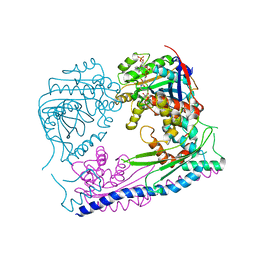 | | Structure of the Rtt109-AcCoA/Vps75 complex and implications for chaperone-mediated histone acetylation | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, Histone acetyltransferase, ... | | Authors: | Tang, Y, Yuan, H, Meeth, K, Marmorstein, R. | | Deposit date: | 2010-12-21 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the Rtt109-AcCoA/Vps75 Complex and Implications for Chaperone-Mediated Histone Acetylation.
Structure, 19, 2011
|
|
7YPD
 
 | |
1U5D
 
 | | Crystal Structure of the PH domain of SKAP55 | | Descriptor: | SULFATE ION, Src Kinase-associated Phosphoprotein of 55 kDa | | Authors: | Tang, Y, Swanson, K.D, Neel, B.G, Eck, M.J. | | Deposit date: | 2004-07-27 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Dimerization and Phosphoinositide Specificity of the Src Kinase-associated Phosphoproteins SKAP55 and SKAP-Hom
To be Published
|
|
3VLU
 
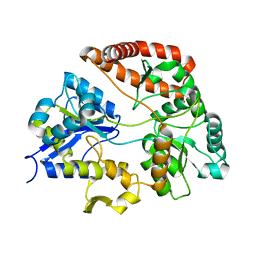 | | Crystal structure of Sphingomonas sp. A1 alginate-binding protein AlgQ1 in complex with saturated trimannuronate | | Descriptor: | AlgQ1, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Nishitani, Y, Maruyama, Y, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Recognition of heteropolysaccharide alginate by periplasmic solute-binding proteins of a bacterial ABC transporter
Biochemistry, 51, 2012
|
|
4YTX
 
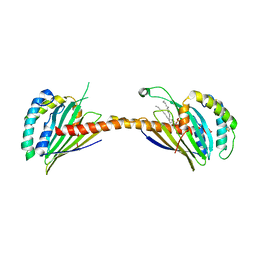 | | Crystal structure of Ups1-Mdm35 complex with PA | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, Mitochondrial distribution and morphology protein 35, Protein UPS1, ... | | Authors: | Watanabe, Y, Tamura, Y, Kawano, S, Endo, T. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria.
Nat Commun, 6, 2015
|
|
4YYH
 
 | | Crystal structure of BRD9 Bromodomain bound to a crotonyllysine peptide | | Descriptor: | Bromodomain-containing protein 9, Histone H4 | | Authors: | Tang, Y, Bellon, S, Cochran, A.G, Poy, F. | | Deposit date: | 2015-03-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications.
Structure, 23, 2015
|
|
3VLV
 
 | | Crystal structure of Sphingomonas sp. A1 alginate-binding ptotein AlgQ1 in complex with unsaturated triguluronate | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, AlgQ1, CALCIUM ION | | Authors: | Nishitani, Y, Maruyama, Y, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of heteropolysaccharide alginate by periplasmic solute-binding proteins of a bacterial ABC transporter
Biochemistry, 51, 2012
|
|
7TAB
 
 | | G-925 bound to the SMARCA4 (BRG1) Bromodomain | | Descriptor: | 2-(6-amino-5-phenylpyridazin-3-yl)phenol, Isoform 4 of Transcription activator BRG1 | | Authors: | Tang, Y, Poy, F, Taylor, A.M, Cochran, A.G, Bellon, S.F. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | GNE-064: A Potent, Selective, and Orally Bioavailable Chemical Probe for the Bromodomains of SMARCA2 and SMARCA4 and the Fifth Bromodomain of PBRM1.
J.Med.Chem., 65, 2022
|
|
7TD9
 
 | | G-059 bound to the SMARCA4 (BRG1) Bromodomain | | Descriptor: | 4-phenyl-5H-pyridazino[4,3-b]indol-3-amine, Isoform 4 of Transcription activator BRG1 | | Authors: | Tang, Y, Poy, F, Taylor, A.M, Cochran, A.G, Bellon, S.F. | | Deposit date: | 2021-12-30 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | GNE-064: A Potent, Selective, and Orally Bioavailable Chemical Probe for the Bromodomains of SMARCA2 and SMARCA4 and the Fifth Bromodomain of PBRM1.
J.Med.Chem., 65, 2022
|
|
3Q33
 
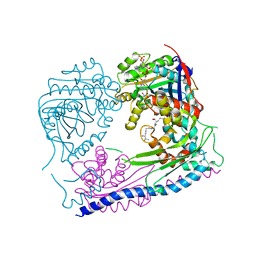 | | Structure of the Rtt109-AcCoA/Vps75 Complex and Implications for Chaperone-Mediated Histone Acetylation | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, HISTONE H3, ... | | Authors: | Tang, Y, Yuan, H, Meeth, K, Marmorstein, R. | | Deposit date: | 2010-12-21 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Rtt109-AcCoA/Vps75 Complex and Implications for Chaperone-Mediated Histone Acetylation.
Structure, 19, 2011
|
|
3VLW
 
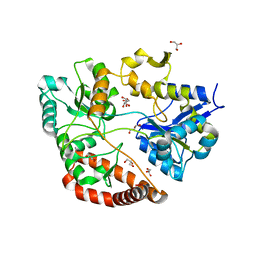 | | Crystal structure of Sphingomonas sp. A1 alginate-binding protein AlgQ1 in complex with mannuronate-guluronate disaccharide | | Descriptor: | AlgQ1, CALCIUM ION, GLYCEROL, ... | | Authors: | Nishitani, Y, Maruyama, Y, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of heteropolysaccharide alginate by periplasmic solute-binding proteins of a bacterial ABC transporter
Biochemistry, 51, 2012
|
|
5AUJ
 
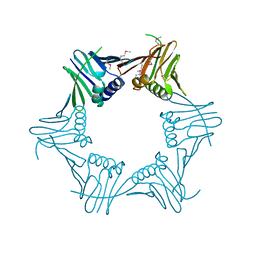 | |
3R7N
 
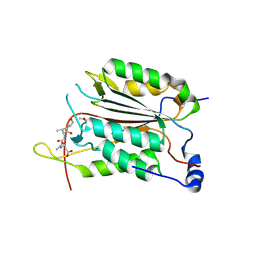 | | Caspase-2 bound with two copies of Ac-DVAD-CHO | | Descriptor: | Caspase-2 subunit p12, Caspase-2 subunit p18, Peptide Inhibitor (ACE)DVAD-CHO | | Authors: | Tang, Y, Wells, J, Arkin, M. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-27 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural and enzymatic insights into caspase-2 protein substrate recognition and catalysis.
J.Biol.Chem., 286, 2011
|
|
6LEP
 
 | | Crystal structure of thiosulfate transporter YeeE inactive mutant - C91A | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Sulf_transp domain-containing protein, THIOSULFATE | | Authors: | Tanaka, Y, Tsukazaki, T, Yoshikaie, K, Sugano, Y, Takeuchi, A, Uchino, S. | | Deposit date: | 2019-11-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a YeeE/YedE family protein engaged in thiosulfate uptake.
Sci Adv, 6, 2020
|
|
6LEO
 
 | | Crystal structure of thiosulfate transporter YeeE from Spirochaeta thermophila | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Sulf_transp domain-containing protein, THIOSULFATE | | Authors: | Tanaka, Y, Tsukazaki, T, Yoshikaie, K, Takeuchi, A, Uchino, S, Sugano, Y. | | Deposit date: | 2019-11-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of a YeeE/YedE family protein engaged in thiosulfate uptake.
Sci Adv, 6, 2020
|
|
5B13
 
 | | Crystal structure of phycoerythrin | | Descriptor: | PHYCOCYANOBILIN, PHYCOUROBILIN, Phycoerythrin alpha subunit, ... | | Authors: | Tanaka, Y, Gai, Z, Kishimura, H. | | Deposit date: | 2015-11-18 | | Release date: | 2016-10-05 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Structural properties of phycoerythrin from dulse palmaria palmata
J FOOD BIOCHEM., 2016
|
|
5I1Q
 
 | | Second bromodomain of TAF1 bound to a pyrrolopyridone compound | | Descriptor: | 3-[6-(but-3-en-1-yl)-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl]-N,N-dimethylbenzamide, Transcription initiation factor TFIID subunit 1 | | Authors: | Tang, Y, Poy, F, Bellon, S.F. | | Deposit date: | 2016-01-09 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diving into the Water: Inducible Binding Conformations for BRD4, TAF1(2), BRD9, and CECR2 Bromodomains.
J.Med.Chem., 59, 2016
|
|
3N9K
 
 | | F229A/E292S Double Mutant of Exo-beta-1,3-glucanase from Candida albicans in Complex with Laminaritriose at 1.7 A | | Descriptor: | CALCIUM ION, Glucan 1,3-beta-glucosidase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Nakatani, Y, Cutfield, S.M, Cutfield, J.F. | | Deposit date: | 2010-05-30 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Carbohydrate binding sites in Candida albicans exo-beta-1,3-glucanase and the role of the Phe-Phe 'clamp' at the active site entrance
Febs J., 277, 2010
|
|
5I29
 
 | | TAF1(2) bound to a pyrrolopyridone compound | | Descriptor: | CALCIUM ION, N,N-dimethyl-3-(6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)benzamide, Transcription initiation factor TFIID subunit 1 | | Authors: | Tang, Y, Poy, F, Bellon, S.F. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Diving into the Water: Inducible Binding Conformations for BRD4, TAF1(2), BRD9, and CECR2 Bromodomains.
J.Med.Chem., 59, 2016
|
|
1C4L
 
 | | SOLUTION STRUCTURE OF AN RNA DUPLEX INCLUDING A C-U BASE-PAIR | | Descriptor: | RNA (5'-R(*CP*CP*UP*GP*CP*GP*UP*CP*G)-3'), RNA (5'-R(*CP*GP*AP*CP*UP*CP*AP*GP*G)-3') | | Authors: | Tanaka, Y, Kojima, C, Yamazaki, T, Kodama, T.S, Yasuno, K, Miyashita, S, Ono, A.M, Ono, A.S, Kainosho, M, Kyogoku, Y. | | Deposit date: | 1999-08-30 | | Release date: | 2000-08-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA duplex including a C-U base pair.
Biochemistry, 39, 2000
|
|
2HG4
 
 | | Structure of the ketosynthase-acyltransferase didomain of module 5 from DEBS. | | Descriptor: | 6-Deoxyerythronolide B Synthase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Tang, Y, Kim, C.Y, Mathews, I.I, Cane, D.E, Khosla, C. | | Deposit date: | 2006-06-26 | | Release date: | 2006-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The 2.7-A crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
