2ZQ5
 
 | |
2ZHH
 
 | | Crystal structure of SoxR | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, FE2/S2 (INORGANIC) CLUSTER, Redox-sensitive transcriptional activator soxR | | Authors: | Watanabe, S, Kita, A, Kobayashi, K, Miki, K. | | Deposit date: | 2008-02-05 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3A43
 
 | | Crystal structure of HypA | | Descriptor: | Hydrogenase nickel incorporation protein hypA, ZINC ION | | Authors: | Watanabe, S, Arai, T, Matsumi, R, Aromi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-06-30 | | Release date: | 2009-10-06 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of HypA, a nickel-binding metallochaperone for [NiFe] hydrogenase maturation.
J.Mol.Biol., 394, 2009
|
|
5FD7
 
 | | X-ray Crystal Structure of ESCRT-III Snf7 core domain (conformation A) | | Descriptor: | Vacuolar-sorting protein SNF7 | | Authors: | Tang, S, Henne, W.M, Borbat, P.P, Buchkovich, N.J, Freed, J.H, Mao, Y, Fromme, J.C, Emr, S.D. | | Deposit date: | 2015-12-15 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments.
Elife, 4, 2015
|
|
5FD9
 
 | | X-ray Crystal Structure of ESCRT-III Snf7 core domain (conformation B) | | Descriptor: | Vacuolar-sorting protein SNF7 | | Authors: | Tang, S, Henne, W.M, Borbat, P.P, Buchkovich, N.J, Freed, J.H, Mao, Y, Fromme, J.C, Emr, S.D. | | Deposit date: | 2015-12-15 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments.
Elife, 4, 2015
|
|
5XWM
 
 | | human ERp44 zinc-bound form | | Descriptor: | CHLORIDE ION, Endoplasmic reticulum resident protein 44, ZINC ION | | Authors: | Watanabe, S, Harayama, M, Inaba, K. | | Deposit date: | 2017-06-30 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Zinc regulates ERp44-dependent protein quality control in the early secretory pathway.
Nat Commun, 10, 2019
|
|
1XUO
 
 | | X-ray structure of LFA-1 I-domain bound to a 1,4-diazepane-2,5-dione inhibitor at 1.8A resolution | | Descriptor: | (2R)-2-[3-ISOBUTYL-2,5-DIOXO-4-(QUINOLIN-3-YLMETHYL)-1,4-DIAZEPAN-1-YL]-N-METHYL-3-(2-NAPHTHYL)PROPANAMIDE, Integrin alpha-L, MAGNESIUM ION | | Authors: | Wattanasin, S, Kallen, J, Myers, S, Guo, Q, Sabio, M, Ehrhardt, C, Albert, R, Hommel, U, Weckbecker, G, Welzenbach, K. | | Deposit date: | 2004-10-26 | | Release date: | 2005-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1,4-Diazepane-2,5-diones as novel inhibitors of LFA-1
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2AW9
 
 | |
1U7Z
 
 | | Phosphopantothenoylcysteine synthetase from E. coli, 4'-phosphopantothenoyl-CMP complex | | Descriptor: | Coenzyme A biosynthesis bifunctional protein coaBC, PHOSPHORIC ACID MONO-[3-(3-{[5-(4-AMINO-2-OXO-2H-PYRIMIDIN-1-YL)-3,4- DIHYDROXY-TETRAHYDRO-FURAN-2- YLMETHOXY]-HYDROXY-PHOSPHORYLOXY}-3-OXO-PROPYLCARBAMOYL)-3-HYDROXY-2,2- DIMETHYL-PROPYL] ESTER | | Authors: | Stanitzek, S, Augustin, M.A, Huber, R, Kupke, T, Steinbacher, S. | | Deposit date: | 2004-08-04 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of CTP-Dependent Peptide Bond Formation in Coenzyme A Biosynthesis Catalyzed by Escherichia coli PPC Synthetase
STRUCTURE, 12, 2004
|
|
1U7U
 
 | | Phosphopantothenoylcysteine synthetase from E. coli | | Descriptor: | Coenzyme A biosynthesis bifunctional protein coaBC | | Authors: | Stanitzek, S, Augustin, M.A, Huber, R, Kupke, T, Steinbacher, S. | | Deposit date: | 2004-08-04 | | Release date: | 2004-11-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of CTP-Dependent Peptide Bond Formation in Coenzyme A Biosynthesis Catalyzed by Escherichia coli PPC Synthetase
STRUCTURE, 12, 2004
|
|
1U80
 
 | | Phosphopantothenoylcysteine synthetase from E. coli, CMP complex | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Coenzyme A biosynthesis bifunctional protein coaBC, PHOSPHATE ION | | Authors: | Stanitzek, S, Augustin, M.A, Huber, R, Kupke, T, Steinbacher, S. | | Deposit date: | 2004-08-04 | | Release date: | 2004-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis of CTP-Dependent Peptide Bond Formation in Coenzyme A Biosynthesis Catalyzed by Escherichia coli PPC Synthetase
STRUCTURE, 12, 2004
|
|
2QW7
 
 | | Carboxysome Subunit, CcmL | | Descriptor: | Carbon dioxide concentrating mechanism protein ccmL, GLYCEROL | | Authors: | Tanaka, S, Sawaya, M.R, Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 2007-08-09 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic-level models of the bacterial carboxysome shell.
Science, 319, 2008
|
|
5AUQ
 
 | | Crystal structure of ATPase-type HypB in the nucleotide free state | | Descriptor: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.525 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AUO
 
 | | Crystal structure of the HypAB-Ni complex (AMPPCP) | | Descriptor: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AUP
 
 | | Crystal structure of the HypAB complex | | Descriptor: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7JST
 
 | | Crystal structure of SARS-CoV-2 3CL in apo form | | Descriptor: | 3C-like proteinase, PHOSPHATE ION | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Huang, Y, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-16 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
7JT0
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with MAC5576 | | Descriptor: | 3C-like proteinase, PHOSPHATE ION, thiophene-2-carbaldehyde | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Huang, Y, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-16 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
7JW8
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with compound 4 in space group P1 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase, ethyl (4R)-4-[[(2S)-4-methyl-2-[[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-2-(phenylmethoxycarbonylamino)butanoyl]amino]pentanoyl]amino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-25 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
7JSU
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with GC376 | | Descriptor: | 3C-like proteinase, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, PHOSPHATE ION | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-16 | | Release date: | 2021-03-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
7JT7
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with compound 4 | | Descriptor: | 3C-like proteinase, ethyl (4R)-4-[[(2S)-4-methyl-2-[[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-2-(phenylmethoxycarbonylamino)butanoyl]amino]pentanoyl]amino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Huang, Y, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-17 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
5AUN
 
 | | Crystal structure of the HypAB-Ni complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase involved in chromosome partitioning, ParA/MinD family, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1U7W
 
 | | Phosphopantothenoylcysteine synthetase from E. coli, CTP-complex | | Descriptor: | CALCIUM ION, CYTIDINE-5'-TRIPHOSPHATE, Coenzyme A biosynthesis bifunctional protein coaBC | | Authors: | Stanitzek, S, Augustin, M.A, Huber, R, Kupke, T, Steinbacher, S. | | Deposit date: | 2004-08-04 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of CTP-Dependent Peptide Bond Formation in Coenzyme A Biosynthesis Catalyzed by Escherichia coli PPC Synthetase
STRUCTURE, 12, 2004
|
|
2VXQ
 
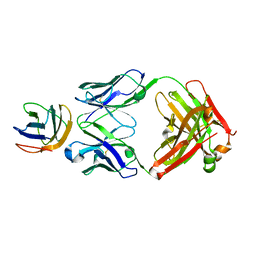 | | Crystal structure of the major grass pollen allergen Phl p 2 in complex with its specific IgE-Fab | | Descriptor: | FAB, POLLEN ALLERGEN PHL P 2 | | Authors: | Padavattan, S, Flicker, S, Schirmer, T, Madritsch, C, Randow, S, Reese, G, Vieths, S, Lupinek, C, Ebner, C, Valenta, R, Markovic-Housley, Z. | | Deposit date: | 2008-07-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-Affinity Ige Recognition of a Conformational Epitope of the Major Respiratory Allergen Phl P 2 as Revealed by X-Ray Crystallography.
J.Immunol., 182, 2009
|
|
2PY8
 
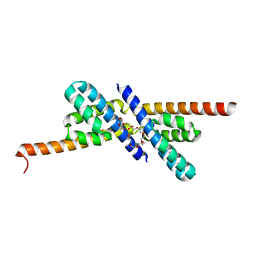 | | RbcX | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CHLORIDE ION, Hypothetical protein rbcX | | Authors: | Tanaka, S, Sawaya, M.R, Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 2007-05-15 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the RuBisCO chaperone RbcX from Synechocystis sp. PCC6803.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2J88
 
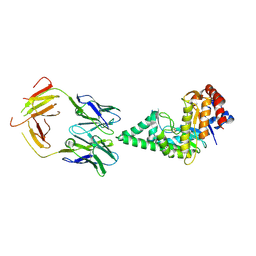 | | Hyaluronidase in complex with a monoclonal IgG Fab fragment | | Descriptor: | FAB, HYALURONONGLUCOSAMINIDASE | | Authors: | Padavattan, S, Schirmer, T, Markovic-Housley, Z. | | Deposit date: | 2006-10-23 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a B-Cell Epitope of Hyaluronidase, a Major Bee Venom Allergen, from its Crystal Structure in Complex with a Specific Fab.
J.Mol.Biol., 368, 2007
|
|
