5XJ4
 
 | | Complex structure of durvalumab-scFv/PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1, durvalumab-VH, durvalumab-VL | | Authors: | Tan, S, Liu, K, Chai, Y, Gao, G.F, Qi, J. | | Deposit date: | 2017-04-29 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Distinct PD-L1 binding characteristics of therapeutic monoclonal antibody durvalumab
Protein Cell, 9, 2018
|
|
8HIT
 
 | | Crystal structure of anti-CTLA-4 humanized IgG1 MAb--JS007 in complex with human CTLA-4 | | Descriptor: | Cytotoxic T-lymphocyte protein 4, JS007-VH, JS007-VL | | Authors: | Tan, S, Shi, Y, Wang, Q, Gao, G.F, Guan, J, Chai, Y, Qi, J. | | Deposit date: | 2022-11-21 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Characterization of the high-affinity anti-CTLA-4 monoclonal antibody JS007 for immune checkpoint therapy of cancer.
Mabs, 15, 2023
|
|
1MNM
 
 | |
1YTF
 
 | | YEAST TFIIA/TBP/DNA COMPLEX | | Descriptor: | DNA (5'-D(*GP*TP*TP*TP*TP*AP*TP*AP*TP*AP*CP*AP*TP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*AP*TP*GP*TP*AP*TP*AP*TP*AP*AP*AP*AP*C)-3'), PROTEIN (TATA BINDING PROTEIN (TBP)), ... | | Authors: | Tan, S, Hunziker, Y, Sargent, D.F, Richmond, T.J. | | Deposit date: | 1996-04-05 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a yeast TFIIA/TBP/DNA complex.
Nature, 381, 1996
|
|
5WT9
 
 | | Complex structure of PD-1 and nivolumab-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Nivolumab, Light Chain of Nivolumab, ... | | Authors: | Tan, S, Zhang, H, Chai, Y, Song, H, Tong, Z, Wang, Q, Qi, J, Wong, G, Zhu, X, Liu, W.J, Gao, S, Wang, Z, Shi, Y, Yang, F, Gao, G.F, Yan, J. | | Deposit date: | 2016-12-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | An unexpected N-terminal loop in PD-1 dominates binding by nivolumab.
Nat Commun, 8, 2017
|
|
9MYM
 
 | | Fertilization IZUMO1 Protein Ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tang, S. | | Deposit date: | 2025-01-22 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.843 Å) | | Cite: | Allosteric inhibition of the IZUMO1-JUNO fertilization complex by the naturally occurring antisperm antibody OBF13.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9MYL
 
 | | Fertilization IZUMO1 Protein Ectodomain in Complex with Anti-sperm Antibody OBF13 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-sperm antibody OBF13 heavy chain, Anti-sperm antibody OBF13 light chain, ... | | Authors: | Tang, S. | | Deposit date: | 2025-01-22 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.177 Å) | | Cite: | Allosteric inhibition of the IZUMO1-JUNO fertilization complex by the naturally occurring antisperm antibody OBF13.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
6IGG
 
 | | Crystal structure of FT condition 1 | | Descriptor: | 1,2-ETHANEDIOL, Protein FLOWERING LOCUS T | | Authors: | Watanabe, S, Nakamura, Y, Kanehara, K, Inaba, K. | | Deposit date: | 2018-09-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-Resolution Crystal Structure of Arabidopsis FLOWERING LOCUS T Illuminates Its Phospholipid-Binding Site in Flowering.
Iscience, 21, 2019
|
|
6IGJ
 
 | | Crystal structure of FT condition 4 | | Descriptor: | MAGNESIUM ION, Protein FLOWERING LOCUS T | | Authors: | Watanabe, S, Nakamura, Y, Kanehara, K, Inaba, K. | | Deposit date: | 2018-09-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | High-Resolution Crystal Structure of Arabidopsis FLOWERING LOCUS T Illuminates Its Phospholipid-Binding Site in Flowering.
Iscience, 21, 2019
|
|
3VYT
 
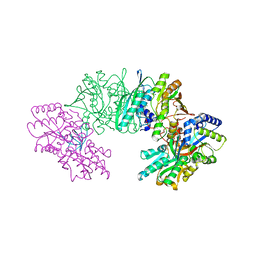 | | Crystal structure of the HypC-HypD-HypE complex (form I inward) | | Descriptor: | CHLORIDE ION, Hydrogenase expression/formation protein HypC, Hydrogenase expression/formation protein HypD, ... | | Authors: | Watanabe, S, Miki, K. | | Deposit date: | 2012-10-02 | | Release date: | 2012-11-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of the HypCD complex and the HypCDE ternary complex: transient intermediate complexes during [NiFe] hydrogenase maturation
Structure, 20, 2012
|
|
3VYR
 
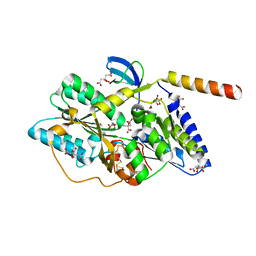 | | Crystal structure of the HypC-HypD complex | | Descriptor: | CITRIC ACID, Hydrogenase expression/formation protein HypC, Hydrogenase expression/formation protein HypD, ... | | Authors: | Watanabe, S, Miki, K. | | Deposit date: | 2012-10-02 | | Release date: | 2012-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of the HypCD complex and the HypCDE ternary complex: transient intermediate complexes during [NiFe] hydrogenase maturation
Structure, 20, 2012
|
|
3VYS
 
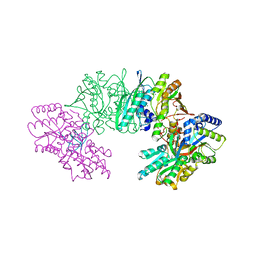 | | Crystal structure of the HypC-HypD-HypE complex (form I) | | Descriptor: | Hydrogenase expression/formation protein HypC, Hydrogenase expression/formation protein HypD, Hydrogenase expression/formation protein HypE, ... | | Authors: | Watanabe, S, Miki, K. | | Deposit date: | 2012-10-02 | | Release date: | 2012-11-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of the HypCD complex and the HypCDE ternary complex: transient intermediate complexes during [NiFe] hydrogenase maturation
Structure, 20, 2012
|
|
3VYU
 
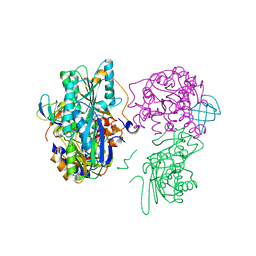 | | Crystal structure of the HypC-HypD-HypE complex (form II) | | Descriptor: | Hydrogenase expression/formation protein HypC, Hydrogenase expression/formation protein HypD, Hydrogenase expression/formation protein HypE, ... | | Authors: | Watanabe, S, Miki, K. | | Deposit date: | 2012-10-02 | | Release date: | 2012-11-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of the HypCD complex and the HypCDE ternary complex: transient intermediate complexes during [NiFe] hydrogenase maturation
Structure, 20, 2012
|
|
6IGI
 
 | | Crystal structure of FT condition 2 | | Descriptor: | 1,2-ETHANEDIOL, Protein FLOWERING LOCUS T | | Authors: | Watanabe, S, Nakamura, Y, Kanehara, K, Inaba, K. | | Deposit date: | 2018-09-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | High-Resolution Crystal Structure of Arabidopsis FLOWERING LOCUS T Illuminates Its Phospholipid-Binding Site in Flowering.
Iscience, 21, 2019
|
|
6IGH
 
 | | Crystal structure of FT condition3 | | Descriptor: | 1,2-ETHANEDIOL, Protein FLOWERING LOCUS T | | Authors: | Watanabe, S, Nakamura, Y, Kanehara, K, Inaba, K. | | Deposit date: | 2018-09-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | High-Resolution Crystal Structure of Arabidopsis FLOWERING LOCUS T Illuminates Its Phospholipid-Binding Site in Flowering.
Iscience, 21, 2019
|
|
2Z1C
 
 | | Crystal structure of HypC from Thermococcus kodakaraensis KOD1 | | Descriptor: | GLYCEROL, Hydrogenase expression/formation protein HypC, TETRAETHYLENE GLYCOL | | Authors: | Watanabe, S, Matsumi, R, Arai, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2007-05-08 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of [NiFe] Hydrogenase Maturation Proteins HypC, HypD, and HypE: Insights into Cyanation Reaction by Thiol Redox Signaling
Mol.Cell, 27, 2007
|
|
2Z9H
 
 | | Ethanolamine utilization protein, EutN | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CHLORIDE ION, Ethanolamine utilization protein eutN | | Authors: | Tanaka, S, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2007-09-20 | | Release date: | 2007-10-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The crystal structure of ethanolamine utilization protein EutN from E. coli
To be Published
|
|
2Z1F
 
 | | Crystal structure of HypE from Thermococcus kodakaraensis (inward form) | | Descriptor: | Hydrogenase expression/formation protein HypE | | Authors: | Watanabe, S, Matsumi, R, Arai, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2007-05-08 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of [NiFe] Hydrogenase Maturation Proteins HypC, HypD, and HypE: Insights into Cyanation Reaction by Thiol Redox Signaling
Mol.Cell, 27, 2007
|
|
2Z1E
 
 | | Crystal structure of HypE from Thermococcus kodakaraensis (outward form) | | Descriptor: | Hydrogenase expression/formation protein HypE | | Authors: | Watanabe, S, Matsumi, R, Arai, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2007-05-08 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structures of [NiFe] Hydrogenase Maturation Proteins HypC, HypD, and HypE: Insights into Cyanation Reaction by Thiol Redox Signaling
Mol.Cell, 27, 2007
|
|
2ZHH
 
 | | Crystal structure of SoxR | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, FE2/S2 (INORGANIC) CLUSTER, Redox-sensitive transcriptional activator soxR | | Authors: | Watanabe, S, Kita, A, Kobayashi, K, Miki, K. | | Deposit date: | 2008-02-05 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7UX0
 
 | | Human Sperm TMEM95 Ectodomain | | Descriptor: | Sperm-egg fusion protein TMEM95, TRIETHYLENE GLYCOL, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Tang, S, Kim, P.S. | | Deposit date: | 2022-05-04 | | Release date: | 2022-10-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Human sperm TMEM95 binds eggs and facilitates membrane fusion.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2ZQ5
 
 | |
2ZHG
 
 | | Crystal structure of SoxR in complex with DNA | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA (5'-D(*DGP*DCP*DCP*DTP*DCP*DAP*DAP*DGP*DTP*DTP*DAP*DAP*DCP*DTP*DTP*DGP*DAP*DGP*DGP*DC)-3'), FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Watanabe, S, Kita, A, Kobayashi, K, Miki, K. | | Deposit date: | 2008-02-05 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2AW9
 
 | |
2D2E
 
 | | Crystal structure of atypical cytoplasmic ABC-ATPase SufC from Thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, GLYCEROL, SufC protein | | Authors: | Watanabe, S, Kita, A, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-09-08 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Atypical Cytoplasmic ABC-ATPase SufC from Thermus thermophilus HB8.
J.Mol.Biol., 353, 2005
|
|
