5XXL
 
 | | Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | Authors: | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahash, Y, Sugimoto, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-07-04 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
5XXO
 
 | | Crystal structure of mutant (D286N) GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorotriose | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | Authors: | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahash, Y, Sugimoto, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-07-04 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
5XXN
 
 | | Crystal Structure of mutant (D286N) beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorose | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | Authors: | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahashi, Y, Sugimono, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-07-04 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
5YP4
 
 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with Lys-Pro from Pseudoxanthomonas mexicana WO24 | | Descriptor: | Dipeptidyl aminopeptidase 4, GLYCEROL, LYSINE, ... | | Authors: | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
5XXM
 
 | | Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with gluconolactone | | Descriptor: | D-glucono-1,5-lactone, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | Authors: | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahash, Y, Sugimoto, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-07-04 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
5YP1
 
 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) from Pseudoxanthomonas mexicana WO24 | | Descriptor: | Dipeptidyl aminopeptidase 4, GLYCEROL | | Authors: | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
5YP2
 
 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with DPP4 inhibitor from Pseudoxanthomonas mexicana WO24 | | Descriptor: | (2S,5R)-1-[2-[[1-(hydroxymethyl)cyclopentyl]amino]ethanoyl]pyrrolidine-2,5-dicarbonitrile, Dipeptidyl aminopeptidase 4, GLYCEROL | | Authors: | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
5YP3
 
 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with Ile-Pro from Pseudoxanthomonas mexicana | | Descriptor: | Dipeptidyl aminopeptidase 4, GLYCEROL, ISOLEUCINE, ... | | Authors: | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
5Z06
 
 | | Crystal structure of beta-1,2-glucanase from Parabacteroides distasonis | | Descriptor: | BDI_3064 protein, CALCIUM ION, GLYCEROL | | Authors: | Shimizu, H, Nakajima, M, Miyanaga, A, Takahashi, Y, Tanaka, N, Kobayashi, K, Sugimoto, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization and Structural Analysis of a Novel exo-Type Enzyme Acting on beta-1,2-Glucooligosaccharides from Parabacteroides distasonis
Biochemistry, 57, 2018
|
|
2ZDS
 
 | | Crystal Structure of SCO6571 from Streptomyces coelicolor A3(2) | | Descriptor: | Putative DNA-binding protein | | Authors: | Begum, P, Gao, Y.G, Sakai, N, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2007-11-27 | | Release date: | 2008-12-02 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of SCO6571 from Streptomyces coelicolor A3(2).
Protein Pept.Lett., 15, 2008
|
|
2Z2E
 
 | | Crystal Structure of Canine Milk Lysozyme Stabilized against Non-enzymatic Deamidation | | Descriptor: | Lysozyme C, milk isozyme, SULFATE ION | | Authors: | Nonaka, Y, Akieda, D, Watanabe, N, Tanaka, I, Kamiya, M, Aizawa, T, Nitta, K, Demura, M, Kawano, K. | | Deposit date: | 2007-05-21 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Spontaneous asparaginyl deamidation of canine milk lysozyme under mild conditions
Proteins, 72, 2008
|
|
8CAT
 
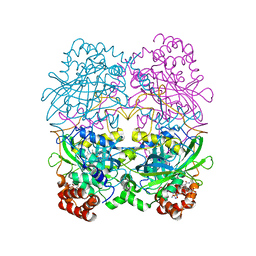 | | The NADPH binding site on beef liver catalase | | Descriptor: | CATALASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murthy, M.R.N, Reid III, T.J, Sicignano, A, Tanaka, N, Fita, I, Rossmann, M.G. | | Deposit date: | 1984-11-15 | | Release date: | 1985-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The NADPH binding site on beef liver catalase.
Proc.Natl.Acad.Sci.USA, 82, 1985
|
|
2D5K
 
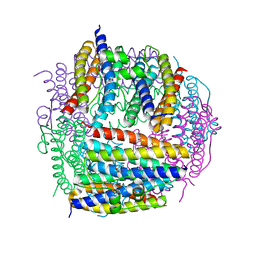 | | Crystal structure of Dps from Staphylococcus aureus | | Descriptor: | Dps family protein, GLYCEROL | | Authors: | Tanaka, Y, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2005-11-02 | | Release date: | 2006-10-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Nucleoid compaction by MrgA(Asp56Ala/Glu60Ala) does not contribute to staphylococcal cell survival against oxidative stress and phagocytic killing by macrophages
FEMS Microbiol. Lett., 360, 2014
|
|
1WU7
 
 | | Crystal structure of histidyl-tRNA synthetase from Thermoplasma acidophilum | | Descriptor: | Histidyl-tRNA synthetase | | Authors: | Tanaka, Y, Sakai, N, Yao, M, Watanabe, N, Tamura, T, Tanaka, I. | | Deposit date: | 2004-12-01 | | Release date: | 2005-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of histidyl-tRNA synthetase from Thermoplasma acidophilum
To be Published
|
|
2Z2F
 
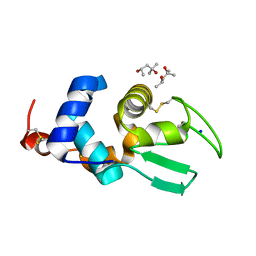 | | X-ray Crystal Structure of Bovine Stomach Lysozyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lysozyme C-2, SODIUM ION | | Authors: | Akieda, D, Nonaka, Y, Watanabe, N, Tanaka, I, Kamiya, M, Aizawa, T, Nitta, K, Demura, M, Kawano, K. | | Deposit date: | 2007-05-21 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Stability of Bovine Stomach Lysozyme in Acidic Condition
To be Published
|
|
1XAB
 
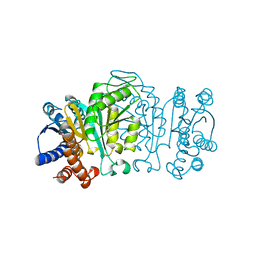 | |
1XAA
 
 | |
2ZOS
 
 | |
2ZU8
 
 | |
2ZOY
 
 | | The multi-drug binding transcriptional repressor CgmR (CGL2612 protein) from C.glutamicum | | Descriptor: | GLYCEROL, Transcriptional regulator | | Authors: | Itou, H, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2008-06-20 | | Release date: | 2008-07-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The CGL2612 protein from Corynebacterium glutamicum is a drug resistance-related transcriptional repressor: structural and functional analysis of a newly identified transcription factor from genomic DNA analysis
J.Biol.Chem., 280, 2005
|
|
2ZB9
 
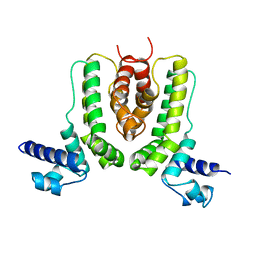 | | Crystal structure of TetR family transcription regulator SCO0332 | | Descriptor: | Putative transcriptional regulator | | Authors: | Okada, U, Kondo, K, Watanabe, N, Yao, M, Tanaka, I. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional analysis of the TetR-family transcriptional regulator SCO0332 from Streptomyces coelicolor
ACTA CRYSTALLOGR.,SECT.D, 64, 2008
|
|
2ZU9
 
 | |
2ZU7
 
 | |
2ZCX
 
 | |
1XER
 
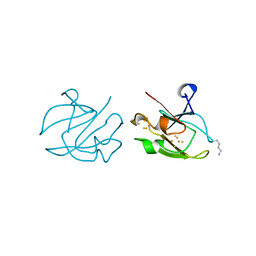 | | STRUCTURE OF FERREDOXIN | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN, ZINC ION | | Authors: | Fujii, T, Hata, Y, Moriyama, H, Wakagi, T, Tanaka, N, Oshima, T. | | Deposit date: | 1996-08-28 | | Release date: | 1997-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel zinc-binding centre in thermoacidophilic archaeal ferredoxins.
Nat.Struct.Biol., 3, 1996
|
|
