3I8N
 
 | |
3I8O
 
 | | A domain of a functionally unknown protein from Methanocaldococcus jannaschii DSM 2661. | | 分子名称: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Tan, K, Chhor, G, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-07-09 | | 公開日 | 2009-07-21 | | 最終更新日 | 2019-07-24 | | 実験手法 | X-RAY DIFFRACTION (2.638 Å) | | 主引用文献 | A domain of a functionally unknown protein from Methanocaldococcus jannaschii DSM 2661.
To be Published
|
|
7TVX
 
 | |
7TVS
 
 | | The Crystal Structure of SARS-CoV-2 Omicron Mpro (P132H) in complex with demethylated analog of masitinib | | 分子名称: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N-(4-methyl-3-{[4-(pyridin-3-yl)-1,3-thiazol-2-yl]amino}phenyl)-4-[(piperazin-1-yl)methyl]benzamide | | 著者 | Tan, K, Maltseva, N.I, Endres, M.J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-02-05 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.88612878 Å) | | 主引用文献 | The Crystal Structure of SARS-CoV-2 Omicron Mpro (P132H) in complex with demethylated analog of masitinib
To Be Published
|
|
7TYE
 
 | |
6W4B
 
 | | The crystal structure of Nsp9 RNA binding protein of SARS CoV-2 | | 分子名称: | Non-structural protein 9 | | 著者 | Tan, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-10 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | The crystal structure of Nsp9 replicase protein of COVID-19
To Be Published
|
|
3IC9
 
 | | The structure of dihydrolipoamide dehydrogenase from Colwellia psychrerythraea 34H. | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, SODIUM ION, dihydrolipoamide dehydrogenase | | 著者 | Tan, K, Rakowski, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-07-17 | | 公開日 | 2009-07-28 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The structure of dihydrolipoamide dehydrogenase from Colwellia psychrerythraea 34H.
To be Published
|
|
3G8W
 
 | | Crystal structure of a probable acetyltransferase from Staphylococcus epidermidis ATCC 12228 | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CITRATE ANION, Lactococcal prophage ps3 protein 05 | | 著者 | Tan, K, Sather, A, Marshall, N, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-02-12 | | 公開日 | 2009-03-03 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The crystal structure of a probable acetyltransferase from Staphylococcus epidermidis ATCC 12228.
To be Published
|
|
3K6H
 
 | | Crystal structure of a nitroreductase family protein from Agrobacterium tumefaciens str. C58 | | 分子名称: | FLAVIN MONONUCLEOTIDE, Nitroreductase family protein, SULFATE ION | | 著者 | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-10-08 | | 公開日 | 2009-10-27 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Crystal structure of a nitroreductase family protein from Agrobacterium tumefaciens str. C58
To be Published
|
|
3IVP
 
 | |
3IUV
 
 | | The structure of a member of TetR family (SCO1917) from Streptomyces coelicolor A3 | | 分子名称: | uncharacterized TetR family protein | | 著者 | Tan, K, Cuff, M, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-08-31 | | 公開日 | 2009-09-22 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.554 Å) | | 主引用文献 | The structure of a member of TetR family (SCO1917) from Streptomyces coelicolor A3
To be Published
|
|
3GKU
 
 | | Crystal structure of a probable RNA-binding protein from Clostridium symbiosum ATCC 14940 | | 分子名称: | Probable RNA-binding protein | | 著者 | Tan, K, Keigher, L, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-03-11 | | 公開日 | 2009-03-31 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | The crystal structure of a probable RNA-binding protein from Clostridium symbiosum ATCC 14940
To be Published
|
|
1LSL
 
 | | Crystal Structure of the Thrombospondin-1 Type 1 Repeats | | 分子名称: | Thrombospondin 1, alpha-L-fucopyranose, beta-L-fucopyranose | | 著者 | Tan, K, Duquette, M, Liu, J, Dong, Y, Zhang, R, Joachimiak, A, Lawler, J, Wang, J.-H. | | 登録日 | 2002-05-17 | | 公開日 | 2002-12-18 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the TSP-1 type 1 repeats: a novel
layered fold and its biological implication.
J.Cell Biol., 159, 2002
|
|
1GEQ
 
 | |
1L6Z
 
 | | CRYSTAL STRUCTURE OF MURINE CEACAM1A[1,4]: A CORONAVIRUS RECEPTOR AND CELL ADHESION MOLECULE IN THE CEA FAMILY | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, biliary glycoprotein C | | 著者 | Tan, K, Zelus, B.D, Meijers, R, Liu, J.-H, Bergelson, J.M, Duke, N, Zhang, R, Joachimiak, A, Holmes, K.V, Wang, J.-H. | | 登録日 | 2002-03-14 | | 公開日 | 2002-09-14 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (3.32 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF MURINE sCEACAM1a[1,4]: A CORONAVIRUS RECEPTOR IN THE CEA FAMILY
Embo J., 21, 2002
|
|
5JMU
 
 | | The crystal structure of the catalytic domain of peptidoglycan N-acetylglucosamine deacetylase from Eubacterium rectale ATCC 33656 | | 分子名称: | ACETATE ION, MAGNESIUM ION, Peptidoglycan N-acetylglucosamine deacetylase, ... | | 著者 | Tan, K, Gu, M, Clancy, S, Joachimiak, A. | | 登録日 | 2016-04-29 | | 公開日 | 2016-06-29 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | The crystal structure of the catalytic domain of peptidoglycan N-acetylglucosamine deacetylase from Eubacterium rectale ATCC 33656 (CASP target)
To Be Published
|
|
5KBP
 
 | | The crystal structure of an alpha-mannosidase from Enterococcus faecalis V583 | | 分子名称: | Glycosyl hydrolase, family 38, SULFATE ION | | 著者 | Tan, K, Chhor, G, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-06-03 | | 公開日 | 2016-07-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The crystal structure of an alpha-mannosidase from Enterococcus faecalis V583
To Be Published
|
|
5JMB
 
 | |
8WDU
 
 | | Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by sucrose density | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Antenna complex alpha/beta subunit, ... | | 著者 | Tani, K, Kanno, R, Harada, A, Kobayashi, A, Minamino, A, Nakamura, N, Ji, X.-C, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Iwasaki, K, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | 登録日 | 2023-09-16 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.24 Å) | | 主引用文献 | High-resolution structure and biochemical properties of the LH1-RC photocomplex from the model purple sulfur bacterium, Allochromatium vinosum.
Commun Biol, 7, 2024
|
|
8WDV
 
 | | Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by Ca2+-DEAE | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Antenna complex alpha/beta subunit, ... | | 著者 | Tani, K, Kanno, R, Harada, A, Kobayashi, A, Minamino, A, Nakamura, N, Ji, X.-C, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Iwasaki, K, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | 登録日 | 2023-09-16 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.24 Å) | | 主引用文献 | High-resolution structure and biochemical properties of the LH1-RC photocomplex from the model purple sulfur bacterium, Allochromatium vinosum.
Commun Biol, 7, 2024
|
|
6V6N
 
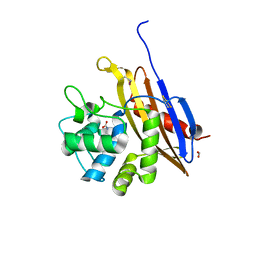 | | The crystal structure of a class D beta-lactamase from Agrobacterium tumefaciens | | 分子名称: | Beta-lactamase, FORMIC ACID, GLYCEROL, ... | | 著者 | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-12-05 | | 公開日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The crystal structure of a class D beta-lactamase from Agrobacterium tumefaciens
To Be Published
|
|
6V4W
 
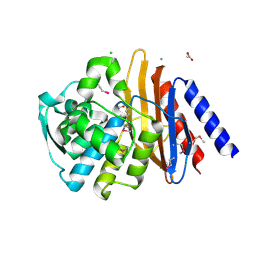 | | The crystal structure of a beta-lactamase from Chitinophaga pinensis DSM 2588 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, Beta-lactamase, ... | | 著者 | Tan, K, Welk, L, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-12-02 | | 公開日 | 2019-12-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | The crystal structure of a beta-lactamase from Chitinophaga pinensis DSM 2588
To Be Published
|
|
6WGQ
 
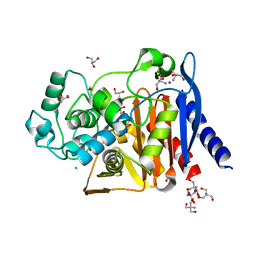 | | The crystal structure of a beta-lactamase from Shigella flexneri 2a str. 2457T | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, CHLORIDE ION, ... | | 著者 | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-06 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The crystal structure of a beta-lactamase from Shigella flexneri 2a str. 2457T
To Be Published
|
|
3L34
 
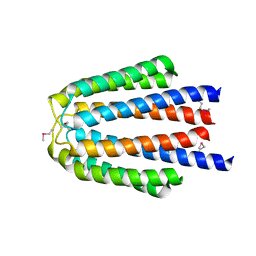 | |
3ON3
 
 | | The crystal structure of keto/oxoacid ferredoxin oxidoreductase, gamma subunit from Geobacter sulfurreducens PCA | | 分子名称: | Keto/oxoacid ferredoxin oxidoreductase, gamma subunit, SULFATE ION | | 著者 | Tan, K, Zhang, R, Hatzos, C, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-08-27 | | 公開日 | 2010-09-22 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.193 Å) | | 主引用文献 | The crystal structure of keto/oxoacid ferredoxin oxidoreductase, gamma subunit from Geobacter sulfurreducens PCA
To be Published
|
|
