6W4B
 
 | | The crystal structure of Nsp9 RNA binding protein of SARS CoV-2 | | Descriptor: | Non-structural protein 9 | | Authors: | Tan, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-10 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The crystal structure of Nsp9 replicase protein of COVID-19
To Be Published
|
|
6X9Y
 
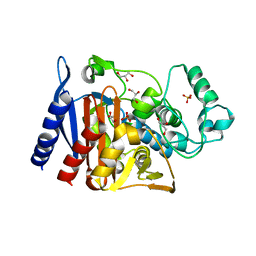 | | The crystal structure of a Beta-lactamase from Escherichia coli CFT073 | | Descriptor: | Beta-lactamase, GLYCEROL, S,R MESO-TARTARIC ACID, ... | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a Beta-lactamase from Escherichia coli CFT073
To Be Published
|
|
3NE8
 
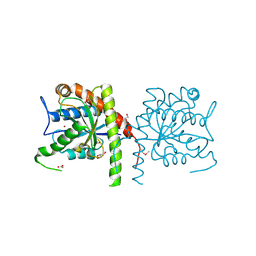 | | The crystal structure of a domain from N-acetylmuramoyl-l-alanine amidase of Bartonella henselae str. Houston-1 | | Descriptor: | ACETATE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Rakowski, E, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-08 | | Release date: | 2010-07-14 | | Last modified: | 2012-11-07 | | Method: | X-RAY DIFFRACTION (1.239 Å) | | Cite: | A conformational switch controls cell wall-remodelling enzymes required for bacterial cell division.
Mol.Microbiol., 85, 2012
|
|
6WHL
 
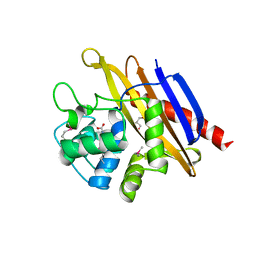 | |
3LHE
 
 | | The crystal structure of the C-terminal domain of a GntR family transcriptional regulator from Bacillus anthracis str. Sterne | | Descriptor: | CHLORIDE ION, GLYCEROL, GntR family Transcriptional regulator | | Authors: | Tan, K, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The crystal structure of the C-terminal domain of a GntR family transcriptional regulator from Bacillus anthracis str. Sterne
To be Published
|
|
3LJL
 
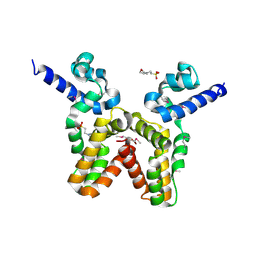 | |
3LHH
 
 | | The crystal structure of CBS domain protein from Shewanella oneidensis MR-1. | | Descriptor: | ADENOSINE MONOPHOSPHATE, CBS domain protein | | Authors: | Tan, K, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of CBS domain protein from Shewanella oneidensis MR-1.
To be Published
|
|
3LOQ
 
 | | The crystal structure of a universal stress protein from Archaeoglobus fulgidus DSM 4304 | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Tan, K, Weger, A, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-04 | | Release date: | 2010-02-16 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The crystal structure of a universal stress protein from Archaeoglobus fulgidus DSM 4304
To be Published
|
|
3LAX
 
 | |
3LAE
 
 | | The crystal structure of a functionally unknown conserved protein from Haemophilus influenzae Rd KW20 | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, UPF0053 protein HI0107, ... | | Authors: | Tan, K, Li, H, Bargassa, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-06 | | Release date: | 2010-01-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.453 Å) | | Cite: | The crystal structure of a functionally unknown conserved protein from Haemophilus influenzae Rd KW20
To be Published
|
|
3LAZ
 
 | |
3LZK
 
 | | The crystal structure of a probably aromatic amino acid degradation proteiN from Sinorhizobium meliloti 1021 | | Descriptor: | CALCIUM ION, Fumarylacetoacetate hydrolase family protein | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-01 | | Release date: | 2010-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a probably aromatic amino acid degradation protein from Sinorhizobium meliloti 1021
To be Published
|
|
3NUK
 
 | | THE CRYSTAL STRUCTURE OF THE W169Y mutant of ALPHA-GLUCOSIDASE (FAMILY 31) from RUMINOCOCCUS OBEUM ATCC 29174 | | Descriptor: | ALPHA-GLUCOSIDASE, GLYCEROL | | Authors: | Tan, K, Tesar, C, Wilton, R, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-07 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | THE CRYSTAL STRUCTURE OF THE W169Y mutant of ALPHA-GLUCOSIDASE (FAMILY 31) from RUMINOCOCCUS OBEUM ATCC 29174
TO BE PUBLISHED
|
|
3NSX
 
 | | The crystal structure of the The crystal structure of the D420A mutant of the alpha-glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, alpha-glucosidase | | Authors: | Tan, K, Tesar, C, Wilton, R, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-02 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.569 Å) | | Cite: | The crystal structure of the The crystal structure of the D420A mutant of the alpha-glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174
To be Published
|
|
3M46
 
 | | The crystal structure of the D73A mutant of glycoside HYDROLASE (FAMILY 31) from Ruminococcus obeum ATCC 29174 | | Descriptor: | GLYCEROL, Uncharacterized protein | | Authors: | Tan, K, Tesar, C, Freeman, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-10 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Novel alpha-glucosidase from human gut microbiome: substrate specificities and their switch
Faseb J., 24, 2010
|
|
3MT0
 
 | | The crystal structure of a functionally unknown protein PA1789 from Pseudomonas aeruginosa PAO1 | | Descriptor: | CHLORIDE ION, uncharacterized protein PA1789 | | Authors: | Tan, K, Chang, C, Tesar, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-29 | | Release date: | 2010-05-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.582 Å) | | Cite: | The crystal structure of a functionally unknown protein PA1789 from Pseudomonas aeruginosa PAO1
To be Published
|
|
3M05
 
 | |
3MUQ
 
 | |
3MO4
 
 | | The crystal structure of an alpha-(1-3,4)-fucosidase from Bifidobacterium longum subsp. infantis ATCC 15697 | | Descriptor: | Alpha-1,3/4-fucosidase, FORMIC ACID, TYROSINE | | Authors: | Tan, K, Xu, X, Cui, H, Ng, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-22 | | Release date: | 2010-05-12 | | Last modified: | 2012-10-10 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Bifidobacterium longum subsp. infantis ATCC 15697 alpha-fucosidases are active on fucosylated human milk oligosaccharides.
Appl.Environ.Microbiol., 78, 2012
|
|
3M33
 
 | | The crystal structure of a functionally unknown protein from Deinococcus radiodurans R1 | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Tan, K, Mack, J, Feldmann, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-08 | | Release date: | 2010-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | The crystal structure of a functionally unknown protein from Deinococcus radiodurans R1
To be Published
|
|
3N04
 
 | | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (FAMILY 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 | | Descriptor: | GLYCEROL, alpha-glucosidase | | Authors: | Tan, K, Tesar, C, Freeman, L, Wilton, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-13 | | Release date: | 2010-06-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (FAMILY 31) FROM RUMINOCOCCUS OBEUM ATCC 29174
Faseb J., 24, 2010
|
|
3NJA
 
 | | The crystal structure of the PAS domain of a GGDEF family protein from Chromobacterium violaceum ATCC 12472. | | Descriptor: | CHLORIDE ION, GLYCEROL, Probable GGDEF family protein, ... | | Authors: | Tan, K, Wu, R, Feldmann, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-08-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.368 Å) | | Cite: | The crystal structure of the PAS domain of a GGDEF family protein from Chromobacterium violaceum ATCC 12472.
To be Published
|
|
3M6D
 
 | | The crystal structure of the d307a mutant of glycoside Hydrolase (family 31) from ruminococcus obeum atcc 29174 | | Descriptor: | Uncharacterized protein | | Authors: | Tan, K, Tesar, C, Freeman, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-15 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Novel alpha-glucosidase from human gut microbiome: substrate specificities and their switch.
Faseb J., 24, 2010
|
|
3OCM
 
 | | The crystal structure of a domain from a possible membrane protein of Bordetella parapertussis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative membrane protein, ... | | Authors: | Tan, K, Tesar, C, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-10 | | Release date: | 2010-10-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The crystal structure of a domain from a possible membrane protein of Bordetella parapertussis
To be Published
|
|
3OCO
 
 | |
