4OG2
 
 | | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with leucine | | Descriptor: | Amino acid/amide ABC transporter substrate-binding protein, HAAT family, CHLORIDE ION, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-15 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.099 Å) | | Cite: | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with leucine
To be Published
|
|
4QN8
 
 | | The crystal structure of an effector protein VipE from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | BETA-MERCAPTOETHANOL, VipE | | Authors: | Tan, K, Xu, X, Cui, H, Liu, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-16 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | The crystal structure of an effector protein VipE from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
To be Published
|
|
4QYM
 
 | | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with methionine | | Descriptor: | Amino acid/amide ABC transporter substrate-binding protein, HAAT family, MAGNESIUM ION, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-24 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with methionine
To be Published
|
|
4NV3
 
 | | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with valine. | | Descriptor: | ACETATE ION, Amino acid/amide ABC transporter substrate-binding protein, HAAT family, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-12-04 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with valine.
To be Published
|
|
3B4S
 
 | |
3B4Q
 
 | |
3BJO
 
 | |
2LM9
 
 | |
2NDK
 
 | |
4ESE
 
 | | The crystal structure of azoreductase from Yersinia pestis CO92 in complex with FMN. | | Descriptor: | DODECAETHYLENE GLYCOL, FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase, ... | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-23 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The crystal structure of acyl carrier protein phosphodiesterase from Yersinia pestis CO92 in complex with FMN.
To be Published
|
|
4DYU
 
 | | The crystal structure of DNA starvation/stationary phase protection protein Dps from Yersinia pestis KIM 10 | | Descriptor: | DNA protection during starvation protein, SULFATE ION, ZINC ION | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-02-29 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The crystal structure of DNA starvation/stationary phase protection protein Dps from Yersinia pestis KIM 10
To be Published
|
|
4HDE
 
 | | The crystal structure of a SCO1/SenC family lipoprotein from Bacillus anthracis str. Ames | | Descriptor: | SCO1/SenC family lipoprotein | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-10-02 | | Release date: | 2012-10-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.317 Å) | | Cite: | The crystal structure of a SCO1/SenC family lipoprotein from Bacillus anthracis str. Ames
To be Published
|
|
2O3F
 
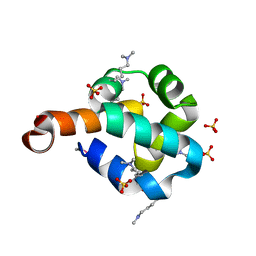 | | Structural Genomics, the crystal structure of the N-terminal domain of the putative transcriptional regulator ybbH from Bacillus subtilis subsp. subtilis str. 168. | | Descriptor: | Putative HTH-type transcriptional regulator ybbH, SULFATE ION | | Authors: | Tan, K, Bigelow, L, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of the N-terminal domain of the putative transcriptional regulator ybbH from Bacillus subtilis subsp. subtilis str. 168.
To be Published
|
|
4FCA
 
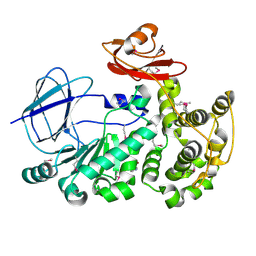 | | The crystal structure of a functionally unknown conserved protein from Bacillus anthracis str. Ames. | | Descriptor: | Conserved domain protein, IMIDAZOLE, NICKEL (II) ION | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-24 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | The crystal structure of a functionally unknown conserved protein from Bacillus anthracis str. Ames.
To be Published
|
|
3UV1
 
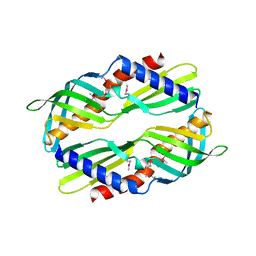 | | Crystal structure a major allergen from dust mite | | Descriptor: | Der f 7 allergen | | Authors: | Tan, K.W, Kumar, T, Chew, F.T, Mok, Y.K. | | Deposit date: | 2011-11-29 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Der f 7, a Dust Mite Allergen from Dermatophagoides farinae.
Plos One, 7, 2012
|
|
4GPN
 
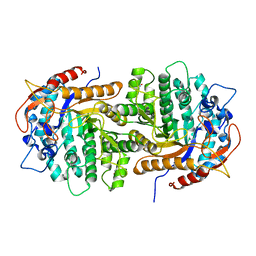 | | The crystal structure of 6-P-beta-D-Glucosidase (E375Q mutant) from Streptococcus mutans UA150 in complex with Gentiobiose 6-phosphate. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-O-phosphono-beta-D-glucopyranose-(1-6)-beta-D-glucopyranose, 6-phospho-beta-D-Glucosidase, ... | | Authors: | Tan, K, Michalska, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
2OUJ
 
 | |
2OR0
 
 | | Structural Genomics, the crystal structure of a putative hydroxylase from Rhodococcus sp. RHA1 | | Descriptor: | ACETATE ION, Hydroxylase | | Authors: | Tan, K, Skarina, T, Kagen, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-01 | | Release date: | 2007-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a putative hydroxylase from Rhodococcus sp. RHA1
To be Published
|
|
4EAE
 
 | | The crystal structure of a functionally unknown protein from Listeria monocytogenes EGD-e | | Descriptor: | D-MALATE, Lmo1068 protein, SODIUM ION | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The crystal structure of a functionally unknown protein from Listeria monocytogenes EGD-e
To be Published
|
|
3UF6
 
 | | The crystal structure of a possible phosphate acetyl/butaryl transferase (from Listeria monocytogenes EGD-e) in complex with CoD (3'-dephosphocoenzyme A) | | Descriptor: | DEPHOSPHO COENZYME A, Lmo1369 protein | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-10-31 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of a possible phosphate acetyl/butaryl transferase (from Listeria monocytogenes EGD-e) in complex with CoD (3'-dephosphocoenzyme A)
To be Published
|
|
4F66
 
 | | The crystal structure of 6-phospho-beta-glucosidase from Streptococcus mutans UA159 in complex with beta-D-glucose-6-phosphate. | | Descriptor: | 1,2-ETHANEDIOL, 6-O-phosphono-beta-D-glucopyranose, FORMIC ACID, ... | | Authors: | Tan, K, Michalska, K, Hatzos-Skintges, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
4F79
 
 | | The crystal structure of 6-phospho-beta-glucosidase mutant (E375Q) in complex with Salicin 6-phosphate | | Descriptor: | 2-(hydroxymethyl)phenyl 6-O-phosphono-beta-D-glucopyranoside, GLYCEROL, Putative phospho-beta-glucosidase | | Authors: | Tan, K, Michalska, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-15 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
6BB9
 
 | | The crystal structure of 4-amino-4-deoxychorismate lyase from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-amino-4-deoxychorismate lyase, ... | | Authors: | Tan, K, Makowska-Grzyska, M, Nocek, B, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-10-17 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | The crystal structure of 4-amino-4-deoxychorismate lyase from Salmonella typhimurium LT2
To Be Published
|
|
3TNG
 
 | | The crystal structure of a possible phosphate acetyl/butaryl transferase from Listeria monocytogenes EGD-e. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lmo1369 protein, NICKEL (II) ION | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-01 | | Release date: | 2011-09-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The crystal structure of a possible phosphate acetyl/butaryl transferase from Listeria monocytogenes EGD-e.
To be Published
|
|
2OUH
 
 | | Crystal structure of the Thrombospondin-1 N-terminal domain in complex with fractionated Heparin DP10 | | Descriptor: | SULFATE ION, Thrombospondin-1 | | Authors: | Tan, K, Joachimiak, A, Wang, J, Lawler, J. | | Deposit date: | 2007-02-11 | | Release date: | 2008-01-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Heparin-induced cis- and trans-Dimerization Modes of the Thrombospondin-1 N-terminal Domain.
J.Biol.Chem., 283, 2008
|
|
