3LAG
 
 | | The crystal structure of a functionally unknown protein RPA4178 from Rhodopseudomonas palustris CGA009 | | 分子名称: | CALCIUM ION, FORMIC ACID, NICKEL (II) ION, ... | | 著者 | Tan, K, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-01-06 | | 公開日 | 2010-01-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | The crystal structure of a functionally unknown protein RPA4178 from Rhodopseudomonas palustris CGA009
To be Published
|
|
3LAZ
 
 | |
3MO4
 
 | | The crystal structure of an alpha-(1-3,4)-fucosidase from Bifidobacterium longum subsp. infantis ATCC 15697 | | 分子名称: | Alpha-1,3/4-fucosidase, FORMIC ACID, TYROSINE | | 著者 | Tan, K, Xu, X, Cui, H, Ng, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-04-22 | | 公開日 | 2010-05-12 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Bifidobacterium longum subsp. infantis ATCC 15697 alpha-fucosidases are active on fucosylated human milk oligosaccharides.
Appl.Environ.Microbiol., 78, 2012
|
|
3MKK
 
 | | The crystal structure of the D307A mutant of glycoside HYDROLASE (FAMILY 31) from Ruminococcus obeum ATCC 29174 in complex with isomaltose | | 分子名称: | alpha-D-glucopyranose, alpha-D-glucopyranose-(1-6)-beta-D-glucopyranose, alpha-glucosidase GH31 family, ... | | 著者 | Tan, K, Tesar, C, Freeman, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-04-15 | | 公開日 | 2010-06-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | The Crystal Structures Of The Glycoside Hydrolase (Family 31) From Ruminococcus Obeum Atcc 29174
Faseb J., 24, 2010
|
|
3MUQ
 
 | |
3NEU
 
 | |
3N04
 
 | | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (FAMILY 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 | | 分子名称: | GLYCEROL, alpha-glucosidase | | 著者 | Tan, K, Tesar, C, Freeman, L, Wilton, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-05-13 | | 公開日 | 2010-06-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (FAMILY 31) FROM RUMINOCOCCUS OBEUM ATCC 29174
Faseb J., 24, 2010
|
|
3M46
 
 | | The crystal structure of the D73A mutant of glycoside HYDROLASE (FAMILY 31) from Ruminococcus obeum ATCC 29174 | | 分子名称: | GLYCEROL, Uncharacterized protein | | 著者 | Tan, K, Tesar, C, Freeman, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-03-10 | | 公開日 | 2010-03-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Novel alpha-glucosidase from human gut microbiome: substrate specificities and their switch
Faseb J., 24, 2010
|
|
3MT0
 
 | | The crystal structure of a functionally unknown protein PA1789 from Pseudomonas aeruginosa PAO1 | | 分子名称: | CHLORIDE ION, uncharacterized protein PA1789 | | 著者 | Tan, K, Chang, C, Tesar, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-04-29 | | 公開日 | 2010-05-12 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.582 Å) | | 主引用文献 | The crystal structure of a functionally unknown protein PA1789 from Pseudomonas aeruginosa PAO1
To be Published
|
|
5JRO
 
 | | The crystal structure of azoreductase from Yersinia pestis CO92 in its Apo form | | 分子名称: | FMN-dependent NADH-azoreductase, GLYCEROL | | 著者 | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-05-06 | | 公開日 | 2016-06-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | The crystal structure of azoreductase from Yersinia pestis CO92 in its Apo form
To Be Published
|
|
3M6D
 
 | | The crystal structure of the d307a mutant of glycoside Hydrolase (family 31) from ruminococcus obeum atcc 29174 | | 分子名称: | Uncharacterized protein | | 著者 | Tan, K, Tesar, C, Freeman, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-03-15 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Novel alpha-glucosidase from human gut microbiome: substrate specificities and their switch.
Faseb J., 24, 2010
|
|
3MZ1
 
 | | The crystal structure of a possible TRANSCRIPTION REGULATOR PROTEIN from Sinorhizobium meliloti 1021 | | 分子名称: | CHLORIDE ION, Putative transcriptional regulator | | 著者 | Tan, K, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-05-11 | | 公開日 | 2010-06-02 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | The crystal structure of a possible TRANSCRIPTION REGULATOR PROTEIN from Sinorhizobium meliloti 1021
To be Published
|
|
3NUK
 
 | | THE CRYSTAL STRUCTURE OF THE W169Y mutant of ALPHA-GLUCOSIDASE (FAMILY 31) from RUMINOCOCCUS OBEUM ATCC 29174 | | 分子名称: | ALPHA-GLUCOSIDASE, GLYCEROL | | 著者 | Tan, K, Tesar, C, Wilton, R, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-07-07 | | 公開日 | 2010-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.055 Å) | | 主引用文献 | THE CRYSTAL STRUCTURE OF THE W169Y mutant of ALPHA-GLUCOSIDASE (FAMILY 31) from RUMINOCOCCUS OBEUM ATCC 29174
TO BE PUBLISHED
|
|
3NSX
 
 | | The crystal structure of the The crystal structure of the D420A mutant of the alpha-glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, alpha-glucosidase | | 著者 | Tan, K, Tesar, C, Wilton, R, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-07-02 | | 公開日 | 2010-07-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.569 Å) | | 主引用文献 | The crystal structure of the The crystal structure of the D420A mutant of the alpha-glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174
To be Published
|
|
5JMU
 
 | | The crystal structure of the catalytic domain of peptidoglycan N-acetylglucosamine deacetylase from Eubacterium rectale ATCC 33656 | | 分子名称: | ACETATE ION, MAGNESIUM ION, Peptidoglycan N-acetylglucosamine deacetylase, ... | | 著者 | Tan, K, Gu, M, Clancy, S, Joachimiak, A. | | 登録日 | 2016-04-29 | | 公開日 | 2016-06-29 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | The crystal structure of the catalytic domain of peptidoglycan N-acetylglucosamine deacetylase from Eubacterium rectale ATCC 33656 (CASP target)
To Be Published
|
|
5KBP
 
 | | The crystal structure of an alpha-mannosidase from Enterococcus faecalis V583 | | 分子名称: | Glycosyl hydrolase, family 38, SULFATE ION | | 著者 | Tan, K, Chhor, G, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-06-03 | | 公開日 | 2016-07-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The crystal structure of an alpha-mannosidase from Enterococcus faecalis V583
To Be Published
|
|
5JMB
 
 | |
6WHL
 
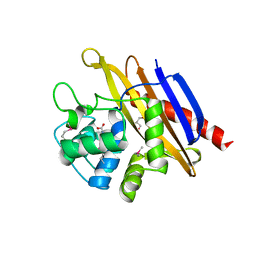 | |
6V6N
 
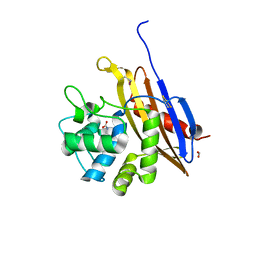 | | The crystal structure of a class D beta-lactamase from Agrobacterium tumefaciens | | 分子名称: | Beta-lactamase, FORMIC ACID, GLYCEROL, ... | | 著者 | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-12-05 | | 公開日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The crystal structure of a class D beta-lactamase from Agrobacterium tumefaciens
To Be Published
|
|
6V4W
 
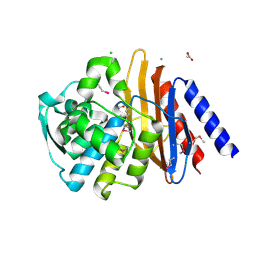 | | The crystal structure of a beta-lactamase from Chitinophaga pinensis DSM 2588 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, Beta-lactamase, ... | | 著者 | Tan, K, Welk, L, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-12-02 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | The crystal structure of a beta-lactamase from Chitinophaga pinensis DSM 2588
To Be Published
|
|
7N6H
 
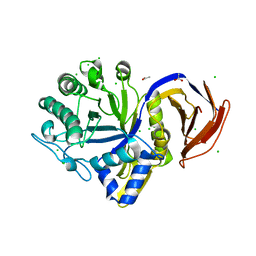 | |
7N6O
 
 | |
6WGQ
 
 | | The crystal structure of a beta-lactamase from Shigella flexneri 2a str. 2457T | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, CHLORIDE ION, ... | | 著者 | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-06 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The crystal structure of a beta-lactamase from Shigella flexneri 2a str. 2457T
To Be Published
|
|
6W4B
 
 | | The crystal structure of Nsp9 RNA binding protein of SARS CoV-2 | | 分子名称: | Non-structural protein 9 | | 著者 | Tan, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-10 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | The crystal structure of Nsp9 replicase protein of COVID-19
To Be Published
|
|
8CZQ
 
 | |
