7F1L
 
 | | Designed enzyme RA61 M48K/I72D mutant: form V | | 分子名称: | CHLORIDE ION, Engineered Retroaldolase, IMIDAZOLE | | 著者 | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | 登録日 | 2021-06-09 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
227D
 
 | | A CRYSTALLOGRAPHIC AND SPECTROSCOPIC STUDY OF THE COMPLEX BETWEEN D(CGCGAATTCGCG)2 AND 2,5-BIS(4-GUANYLPHENYL)FURAN, AN ANALOGUE OF BERENIL. STRUCTURAL ORIGINS OF ENHANCED DNA-BINDING AFFINITY | | 分子名称: | 2,5-BIS(4-GUANYLPHENYL)FURAN, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | 著者 | Laughton, C.A, Tanious, F, Nunn, C.M, Boykin, D.W, Wilson, W.D, Neidle, S. | | 登録日 | 1995-08-08 | | 公開日 | 1995-11-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A crystallographic and spectroscopic study of the complex between d(CGCGAATTCGCG)2 and 2,5-bis(4-guanylphenyl)furan, an analogue of berenil. Structural origins of enhanced DNA-binding affinity.
Biochemistry, 35, 1996
|
|
2WZL
 
 | | The Structure of the N-RNA Binding Domain of the Mokola virus Phosphoprotein | | 分子名称: | GLYCEROL, PHOSPHOPROTEIN | | 著者 | Assenberg, R, Delmas, O, Ren, J, Vidalain, P, Verma, A, Larrous, F, Graham, S, Tangy, F, Grimes, J, Bourhy, H. | | 登録日 | 2009-11-30 | | 公開日 | 2009-12-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Structure of the N-RNA Binding Domain of the Mokola Virus Phosphoprotein
J.Virol., 84, 2010
|
|
3SE5
 
 | | Fic protein from NEISSERIA MENINGITIDIS mutant delta8 in complex with AMPPNP | | 分子名称: | Cell filamentation protein Fic-related protein, HEXAETHYLENE GLYCOL, MAGNESIUM ION, ... | | 著者 | Goepfert, A, Stanger, F, Schirmer, T. | | 登録日 | 2011-06-10 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Adenylylation control by intra- or intermolecular active-site obstruction in Fic proteins.
Nature, 482, 2012
|
|
1KG8
 
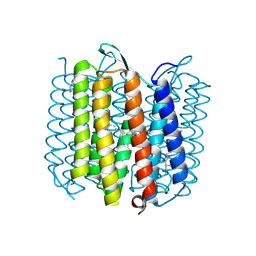 | | X-ray structure of an early-M intermediate of bacteriorhodopsin | | 分子名称: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, RETINAL, bacteriorhodopsin | | 著者 | Facciotti, M.T, Rouhani, S, Burkard, F.T, Betancourt, F.M, Downing, K.H, Rose, R.B, McDermott, G, Glaeser, R.M. | | 登録日 | 2001-11-26 | | 公開日 | 2001-12-05 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of an early intermediate in the M-state phase of the bacteriorhodopsin photocycle.
Biophys.J., 81, 2001
|
|
1ON8
 
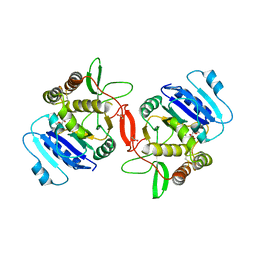 | | Crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) with UDP and GlcUAb(1-3)Galb(1-O)-naphthalenelmethanol an acceptor substrate analog | | 分子名称: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2, MANGANESE (II) ION, ... | | 著者 | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | 登録日 | 2003-02-27 | | 公開日 | 2003-04-22 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
1OMX
 
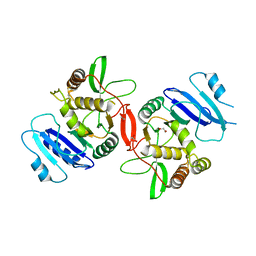 | | Crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) | | 分子名称: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2 | | 著者 | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | 登録日 | 2003-02-26 | | 公開日 | 2003-04-22 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
1OMZ
 
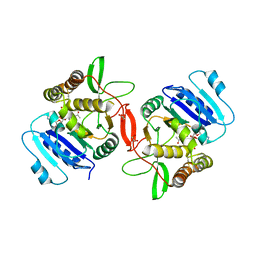 | | crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) in complex with UDPGalNAc | | 分子名称: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2, MANGANESE (II) ION, ... | | 著者 | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | 登録日 | 2003-02-26 | | 公開日 | 2003-04-22 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2),
a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
1KGB
 
 | | structure of ground-state bacteriorhodopsin | | 分子名称: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, RETINAL, bacteriorhodopsin | | 著者 | Facciotti, M.T, Rouhani, S, Burkard, F.T, Betancourt, F.M, Downing, K.H, Rose, R.B, McDermott, G, Glaeser, R.M. | | 登録日 | 2001-11-26 | | 公開日 | 2001-12-05 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structure of an early intermediate in the M-state phase of the bacteriorhodopsin photocycle.
Biophys.J., 81, 2001
|
|
3SN9
 
 | |
1ON6
 
 | | Crystal structure of mouse alpha-1,4-N-acetylhexosaminotransferase (EXTL2) in complex with UDPGlcNAc | | 分子名称: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2, MANGANESE (II) ION, ... | | 著者 | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | 登録日 | 2003-02-27 | | 公開日 | 2003-04-22 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
1KG9
 
 | | Structure of a "mock-trapped" early-M intermediate of bacteriorhosopsin | | 分子名称: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, RETINAL, bacteriorhodopsin | | 著者 | Facciotti, M.T, Rouhani, S, Burkard, F.T, Betancourt, F.M, Downing, K.H, Rose, R.B, McDermott, G, Glaeser, R.M. | | 登録日 | 2001-11-26 | | 公開日 | 2001-12-05 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structure of an early intermediate in the M-state phase of the bacteriorhodopsin photocycle.
Biophys.J., 81, 2001
|
|
1WB7
 
 | | Iron Superoxide Dismutase (Fe-SOD) From The Hyperthermophile Sulfolobus Solfataricus. Crystal Structure of the Y41F mutant. | | 分子名称: | FE (III) ION, SUPEROXIDE DISMUTASE [FE] | | 著者 | Gogliettino, M.A, Tanfani, F, Scire, A, Ursby, T, Adinolfi, B.S, Cacciamani, T, De Vendittis, E. | | 登録日 | 2004-10-31 | | 公開日 | 2004-11-08 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | The Role of Tyr41 and His155 in the Functional Properties of Superoxide Dismutase from the Archaeon Sulfolobus Solfataricus
Biochemistry, 43, 2004
|
|
5YYP
 
 | | Structure K137A thaumatin | | 分子名称: | GLYCEROL, L(+)-TARTARIC ACID, Preprothaumatin I | | 著者 | Masuda, T, Kigo, S, Mitsumoto, M, Ohta, K, Suzuki, M, Mikami, B, Kitabatake, N, Tani, F. | | 登録日 | 2017-12-10 | | 公開日 | 2018-03-21 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.01 Å) | | 主引用文献 | Positive Charges on the Surface of Thaumatin Are Crucial for the Multi-Point Interaction with the Sweet Receptor.
Front Mol Biosci, 5, 2018
|
|
5YYQ
 
 | | Structure K78A thaumatin | | 分子名称: | GLYCEROL, L(+)-TARTARIC ACID, Preprothaumatin I | | 著者 | Masuda, T, Kigo, S, Mitsumoto, M, Ohta, K, Suzuki, M, Mikami, B, Kitabatake, N, Tani, F. | | 登録日 | 2017-12-10 | | 公開日 | 2018-03-21 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.07 Å) | | 主引用文献 | Positive Charges on the Surface of Thaumatin Are Crucial for the Multi-Point Interaction with the Sweet Receptor.
Front Mol Biosci, 5, 2018
|
|
7ZB3
 
 | |
5YYR
 
 | | Structure K106A thaumatin | | 分子名称: | GLYCEROL, L(+)-TARTARIC ACID, Preprothaumatin I | | 著者 | Masuda, T, Kigo, S, Ohta, K, Mitsumoto, M, Mikami, B, Suzuki, M, Kitabatake, N, Tani, F. | | 登録日 | 2017-12-10 | | 公開日 | 2018-03-21 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.07 Å) | | 主引用文献 | Positive Charges on the Surface of Thaumatin Are Crucial for the Multi-Point Interaction with the Sweet Receptor.
Front Mol Biosci, 5, 2018
|
|
3VJQ
 
 | |
3VHG
 
 | |
3WOU
 
 | |
3VHF
 
 | | plant thaumatin I at pH 8.0 | | 分子名称: | GLYCEROL, Thaumatin I | | 著者 | Masuda, T, Mikami, B, Kitabatake, N, Tani, F. | | 登録日 | 2011-08-24 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Atomic structure of the sweet-tasting protein thaumatin I at pH 8.0 reveals the large disulfide-rich region in domain II to be sensitive to a pH change
Biochem.Biophys.Res.Commun., 419, 2012
|
|
