5X6U
 
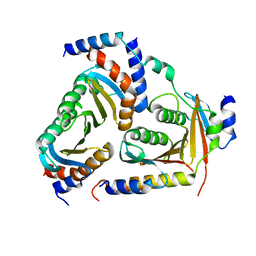 | | Crystal structure of human heteropentameric complex | | Descriptor: | Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, Ragulator complex protein LAMTOR3, ... | | Authors: | Yonehara, R, Nada, S, Nakai, T, Nakai, M, Kitamura, A, Ogawa, A, Nakatsumi, H, Nakayama, K.I, Li, S, Standley, D.M, Yamashita, E, Nakagawa, A, Okada, M. | | Deposit date: | 2017-02-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the assembly of the Ragulator-Rag GTPase complex.
Nat Commun, 8, 2017
|
|
5WEX
 
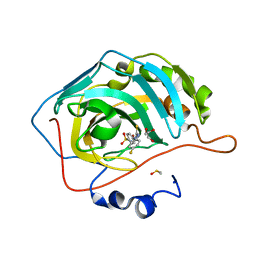 | | Discovery of new selenoureido analogs of 4-(4-fluorophenylureido) benzenesulfonamides as carbonic anhydrase inhibitors | | Descriptor: | 4-{[(4-fluorophenyl)carbamothioyl]amino}benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Angeli, A, Tanini, D, Peat, T.S, Di Cesare Mannelli, L, Bartolucci, G, Capperucci, A, Ghelardini, C, Supuran, C.T, Carta, F. | | Deposit date: | 2017-07-10 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Discovery of New Selenoureido Analogues of 4-(4-Fluorophenylureido)benzenesulfonamide as Carbonic Anhydrase Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
2M1X
 
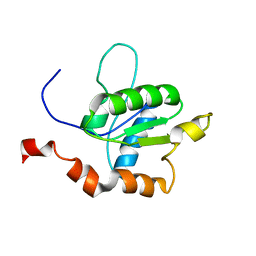 | | TICAM-1 TIR domain structure | | Descriptor: | TIR domain-containing adapter molecule 1 | | Authors: | Enokizono, Y, Kumeta, H, Funami, K, Horiuchi, M, Sarmiento, J, Yamashita, K, Standley, D.M, Matsumoto, M, Seya, T, Inagaki, F. | | Deposit date: | 2012-12-07 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures and interface mapping of the TIR domain-containing adaptor molecules involved in interferon signaling.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5X6V
 
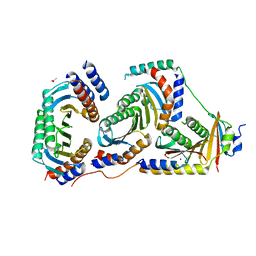 | | Crystal structure of human heteroheptameric complex | | Descriptor: | ACETATE ION, Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, ... | | Authors: | Yonehara, R, Nada, S, Nakai, T, Nakai, M, Kitamura, A, Ogawa, A, Nakatsumi, H, Nakayama, K.I, Li, S, Standley, D.M, Yamashita, E, Nakagawa, A, Okada, M. | | Deposit date: | 2017-02-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for the assembly of the Ragulator-Rag GTPase complex.
Nat Commun, 8, 2017
|
|
6Z9M
 
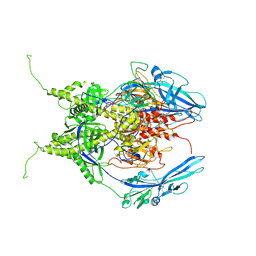 | | Pseudoatomic model of the pre-fusion conformation of glycoprotein B of Herpes simplex virus 1 | | Descriptor: | Envelope glycoprotein B | | Authors: | Vollmer, B, Prazak, V, Vasishtan, D, Jefferys, E.E, Hernandez-Duran, A, Vallbracht, M, Klupp, B, Mettenleiter, T.C, Backovic, M, Rey, F.A, Topf, M, Gruenewald, K. | | Deposit date: | 2020-06-04 | | Release date: | 2020-10-07 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | The prefusion structure of herpes simplex virus glycoprotein B.
Sci Adv, 6, 2020
|
|
2N5J
 
 | | Regnase-1 N-terminal domain | | Descriptor: | Ribonuclease ZC3H12A | | Authors: | Yokogawa, M, Tsushima, T, Noda, N.N, Kumeta, H, Adachi, W, Enokizono, Y, Yamashita, K, Standley, D.M, Takeuchi, O, Akira, S, Inagaki, F. | | Deposit date: | 2015-07-18 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
2N5K
 
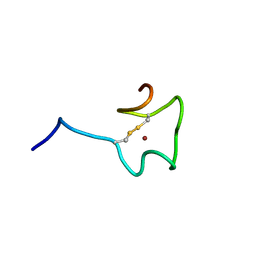 | | Regnase-1 Zinc finger domain | | Descriptor: | Ribonuclease ZC3H12A, ZINC ION | | Authors: | Yokogawa, M, Tsushima, T, Noda, N.N, Kumeta, H, Adachi, W, Enokizono, Y, Yamashita, K, Standley, D.M, Takeuchi, O, Akira, S, Inagaki, F. | | Deposit date: | 2015-07-18 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
4CYL
 
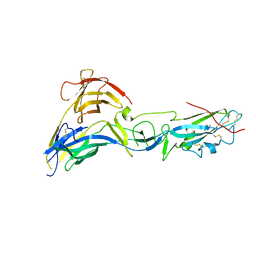 | |
2M1W
 
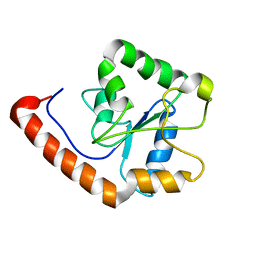 | | TICAM-2 TIR domain | | Descriptor: | TIR domain-containing adapter molecule 2 | | Authors: | Enokizono, Y, Kumeta, H, Funami, K, Horiuchi, M, Sarmiento, J, Yamashita, K, Standley, D.M, Matsumoto, M, Seya, T, Inagaki, F. | | Deposit date: | 2012-12-07 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures and interface mapping of the TIR domain-containing adaptor molecules involved in interferon signaling.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2N5L
 
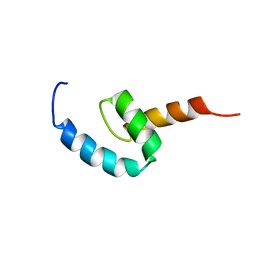 | | Regnase-1 C-terminal domain | | Descriptor: | Ribonuclease ZC3H12A | | Authors: | Yokogawa, M, Tsushima, T, Noda, N.N, Kumeta, H, Adachi, W, Enokizono, Y, Yamashita, K, Standley, D.M, Takeuchi, O, Akira, S, Inagaki, F. | | Deposit date: | 2015-07-18 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
6MJ0
 
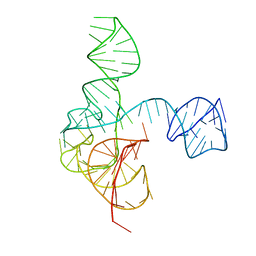 | | Crystal structure of the complete turnip yellow mosaic virus 3'UTR | | Descriptor: | RNA (101-MER) | | Authors: | Hartwick, E.W, Costantino, D.A, MacFadden, A, Nix, J.C, Tian, S, Das, R, Kieft, J.S. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Ribosome-induced RNA conformational changes in a viral 3'-UTR sense and regulate translation levels.
Nat Commun, 9, 2018
|
|
7VEQ
 
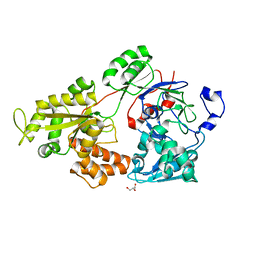 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in an open conformation | | Descriptor: | GLYCEROL, SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VER
 
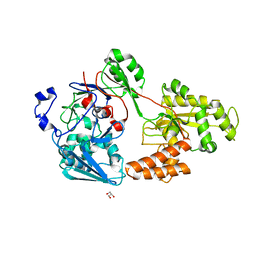 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in a full open conformation | | Descriptor: | GLYCEROL, SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
6D3P
 
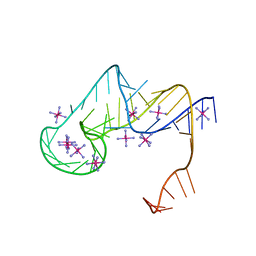 | | Crystal structure of an exoribonuclease-resistant RNA from Sweet clover necrotic mosaic virus (SCNMV) | | Descriptor: | IRIDIUM HEXAMMINE ION, RNA (45-MER) | | Authors: | Steckelberg, A.-L, Akiyama, B.M, Costantino, D.A, Sit, T.L, Nix, J.C, Kieft, J.S. | | Deposit date: | 2018-04-16 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A folded viral noncoding RNA blocks host cell exoribonucleases through a conformationally dynamic RNA structure.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7PTV
 
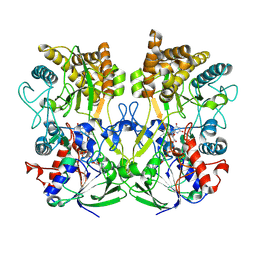 | | Structure of the Mimivirus genomic fibre asymmetric unit | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2021-09-27 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
4PQV
 
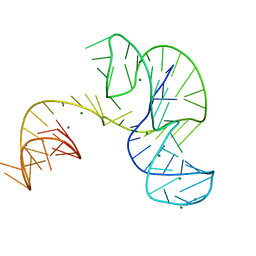 | | Crystal structure of an Xrn1-resistant RNA from the 3' untranslated region of a flavivirus (Murray Valley Encephalitis virus) | | Descriptor: | MAGNESIUM ION, XRN1-resistant flaviviral RNA | | Authors: | Chapman, E.G, Costantino, D.A, Rabe, J.L, Moon, S.L, Wilusz, J, Nix, J.C, Kieft, J.S. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.463 Å) | | Cite: | The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production.
Science, 344, 2014
|
|
7JJU
 
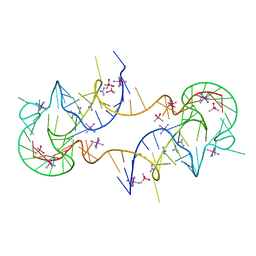 | | Crystal structure of en exoribonuclease-resistant RNA (xrRNA) from Potato leafroll virus (PLRV) | | Descriptor: | CACODYLATE ION, Guanidinium, IRIDIUM HEXAMMINE ION, ... | | Authors: | Steckelberg, A.-L, Vicens, Q, Auffinger, P, Costantino, D.C, Nix, J.C, Kieft, J.S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | The crystal structure of a Polerovirus exoribonuclease-resistant RNA shows how diverse sequences are integrated into a conserved fold.
Rna, 26, 2020
|
|
4P5J
 
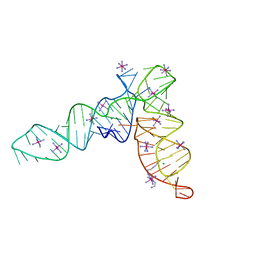 | | Crystal structure of the tRNA-like structure from Turnip Yellow Mosaic Virus (TYMV), a tRNA mimicking RNA | | Descriptor: | IRIDIUM HEXAMMINE ION, MAGNESIUM ION, SPERMINE, ... | | Authors: | Colussi, T.M, Costantino, D.A, Hammond, J.A, Ruehle, G.M, Nix, J.C, Kieft, J.S. | | Deposit date: | 2014-03-17 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9912 Å) | | Cite: | The structural basis of transfer RNA mimicry and conformational plasticity by a viral RNA.
Nature, 511, 2014
|
|
4BKN
 
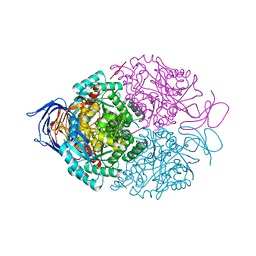 | | Human Dihydropyrimidinase-related protein 3 (DPYSL3) | | Descriptor: | DIHYDROPYRIMIDINASE-RELATED PROTEIN 3 | | Authors: | Mathea, S, Elkins, J.M, Alegre-Abarrategui, J, Shrestha, L, Burgess-Brown, N, Puranik, S, Coutandin, D, Bradley, A, Vollmar, M, von Delft, F, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2013-04-26 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Dpysl3
To be Published
|
|
4BQM
 
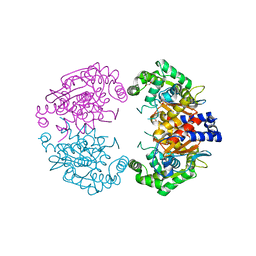 | | Crystal structure of human liver-type glutaminase, catalytic domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLUTAMINASE LIVER ISOFORM, ... | | Authors: | Ferreira, I.M, Vollmar, M, Krojer, T, Strain-Damerell, C, Froese, S, Coutandin, D, Williams, E, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Dias, S.M.G, Ambrosio, A.L.B, Yue, W.W. | | Deposit date: | 2013-05-31 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal Structure of Human Liver-Type Glutaminase, Catalytic Domain
To be Published
|
|
2X80
 
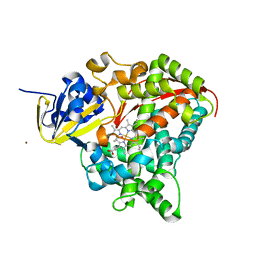 | | P450 BM3 F87A in complex with DMSO | | Descriptor: | BIFUNCTIONAL P-450/NADPH-P450 REDUCTASE, DIMETHYL SULFOXIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kuper, J, Wong, T.S, Roccatano, D, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2010-03-05 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Role of Active-Site Phe87 in Modulating the Organic Co-Solvent Tolerance of Cytochrome P450 Bm3 Monooxygenase.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
7YX4
 
 | | Structure of the Mimivirus genomic fibre in its compact 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
7VET
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in a closed conformation | | Descriptor: | SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEV
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
2X7Y
 
 | | P450 BM3 F87A in complex with DMSO | | Descriptor: | BIFUNCTIONAL P-450/NADPH-P450 REDUCTASE, DIMETHYL SULFOXIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kuper, J, Wong, T.S, Roccatano, D, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2010-03-04 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Role of Active-Site Phe87 in Modulating the Organic Co-Solvent Tolerance of Cytochrome P450 Bm3 Monooxygenase.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
