1UTY
 
 | |
1GWP
 
 | | STRUCTURE OF THE N-TERMINAL DOMAIN OF THE MATURE HIV-1 CAPSID PROTEIN | | Descriptor: | GAG POLYPROTEIN | | Authors: | Tang, C, Gitti, R.K, Lee, B.M, Walker, J, Summers, M.F, Yoo, S, Sundquist, W.I. | | Deposit date: | 2002-03-22 | | Release date: | 2002-06-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-Terminal 283-Residue Fragment of the Immature HIV-1 Gag Polyprotein
Nat.Struct.Biol., 9, 2002
|
|
2W9X
 
 | | The active site of a carbohydrate esterase displays divergent catalytic and non-catalytic binding functions | | Descriptor: | GLYCEROL, PUTATIVE ACETYL XYLAN ESTERASE | | Authors: | Montanier, C, Money, V.A, Pires, V, Flint, J.E, Benedita, P.A, Goyal, A, Prates, J.A, Izumi, A, Stalbrand, H, Morland, C, Cartmell, A, Kolenova, K, Topakas, E, Dobson, E, Bolam, D.N, Davies, G.J, Fontes, C.M, Gilbert, H.J. | | Deposit date: | 2009-01-29 | | Release date: | 2009-03-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Active Site of a Carbohydrate Esterase Displays Divergent Catalytic and Noncatalytic Binding Functions.
Plos Biol., 7, 2009
|
|
2LCU
 
 | | NMR structure of BC28.1 | | Descriptor: | Bc28.1 | | Authors: | Roumestand, C, Delbecq, S, Yang, Y. | | Deposit date: | 2011-05-09 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of Bc28.1, Major Erythrocyte-binding Protein from Babesia canis Merozoite Surface.
J.Biol.Chem., 287, 2012
|
|
2XFE
 
 | | vCBM60 in complex with galactobiose | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING MODULE, beta-D-galactopyranose-(1-4)-beta-D-galactopyranose | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D.P, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules.
J.Biol.Chem., 285, 2010
|
|
2XHJ
 
 | | Circular permutation provides an evolutionary link between two families of calcium-dependent carbohydrate binding modules. SeMet form of vCBM60. | | Descriptor: | CALCIUM ION, CALCIUM-DEPENDENT CARBOHYDRATE BINDING MODULE | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D, Ratnaparkhe, S, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-06-16 | | Release date: | 2010-07-21 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules
J.Biol.Chem., 285, 2010
|
|
2XFD
 
 | | vCBM60 in complex with cellobiose | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING MODULE, GLYCEROL, ... | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D.P, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules.
J.Biol.Chem., 285, 2010
|
|
2XHH
 
 | | Circular permutation provides an evolutionary link between two families of calcium-dependent carbohydrate binding modules | | Descriptor: | (2S)-2-hydroxybutanedioic acid, CALCIUM ION, CARBOHYDRATE BINDING MODULE | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D, Ratnaparkhe, S, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-06-16 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules
J.Biol.Chem., 285, 2010
|
|
7TCZ
 
 | | Human cytomegalovirus protease mutant (C84A, C87A, C138A, C202A) in complex with inhibitor | | Descriptor: | Assemblin, [1-(2-oxopropyl)-4-phenyl-1H-1,2,3-triazol-5-yl]methyl benzylcarbamate | | Authors: | Hulce, K.R, Bohn, M, Ongpipattanakul, C, Jaishankar, P, Renslo, A.R, Craik, C.S. | | Deposit date: | 2021-12-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Inhibiting a dynamic viral protease by targeting a non-catalytic cysteine.
Cell Chem Biol, 29, 2022
|
|
5OR3
 
 | | Crystal structure of Aspergillus oryzae catechol oxidase in met/deoxy-form | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hakulinen, N, Penttinen, L, Rutanen, C, Rouvinen, J. | | Deposit date: | 2017-08-15 | | Release date: | 2018-05-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | A new crystal form of Aspergillus oryzae catechol oxidase and evaluation of copper site structures in coupled binuclear copper enzymes.
PLoS ONE, 13, 2018
|
|
1D5Q
 
 | | SOLUTION STRUCTURE OF A MINI-PROTEIN REPRODUCING THE CORE OF THE CD4 SURFACE INTERACTING WITH THE HIV-1 ENVELOPE GLYCOPROTEIN | | Descriptor: | CHIMERIC MINI-PROTEIN | | Authors: | Vita, C, Drakopoulou, E, Vizzanova, J, Rochette, S, Martin, L, Menez, A, Roumestand, C, Yang, Y.S, Ylisastigui, L, Benjouad, A, Gluckman, J.C. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-11 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Rational engineering of a miniprotein that reproduces the core of the CD4 site interacting with HIV-1 envelope glycoprotein.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1CXN
 
 | | REFINED THREE-DIMENSIONAL SOLUTION STRUCTURE OF A SNAKE CARDIOTOXIN: ANALYSIS OF THE SIDE-CHAIN ORGANISATION SUGGESTS THE EXISTENCE OF A POSSIBLE PHOSPHOLIPID BINDING SITE | | Descriptor: | CARDIOTOXIN GAMMA | | Authors: | Gilquin, B, Roumestand, C, Zinn-Justin, S, Menez, A, Toma, F. | | Deposit date: | 1994-07-08 | | Release date: | 1994-12-20 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Refined three-dimensional solution structure of a snake cardiotoxin: analysis of the side-chain organization suggests the existence of a possible phospholipid binding site.
Biopolymers, 33, 1993
|
|
5XBO
 
 | | Lanthanoid tagging via an unnatural amino acid for protein structure characterization | | Descriptor: | Polyubiquitin-B, TERBIUM(III) ION, UV excision repair protein RAD23 homolog A | | Authors: | Jiang, W, Gu, X, Dong, X, Tang, C. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Lanthanoid tagging via an unnatural amino acid for protein structure characterization
J. Biomol. NMR, 67, 2017
|
|
9F5H
 
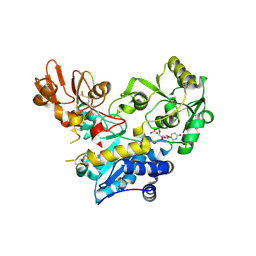 | | Crystal structure of MGAT5 bump-and-hole mutant in complex with UDP and M592 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Secreted alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | Authors: | Liu, Y, Bineva-Todd, G, Meek, R, Mazo, L, Piniello, B, Moroz, O.V, Begum, N, Roustan, C, Tomita, S, Kjaer, S, Rovira, C, Davies, G.J, Schumann, B. | | Deposit date: | 2024-04-28 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A Bioorthogonal Precision Tool for Human N -Acetylglucosaminyltransferase V.
J.Am.Chem.Soc., 146, 2024
|
|
6KOX
 
 | |
6ZUO
 
 | | Human RIO1(kd)-StHA late pre-40S particle, structural state A (pre 18S rRNA cleavage) | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Plassart, L, Shayan, R, Plisson-Chastang, C. | | Deposit date: | 2020-07-23 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The final step of 40S ribosomal subunit maturation is controlled by a dual key lock.
Elife, 10, 2021
|
|
6ZV6
 
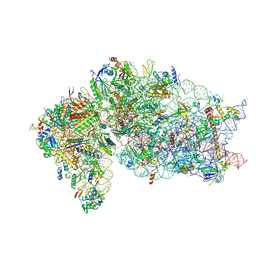 | | Human RIO1(kd)-StHA late pre-40S particle, structural state B (post 18S rRNA cleavage) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Plassart, L, Shayan, R, Plisson-Chastang, C. | | Deposit date: | 2020-07-24 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The final step of 40S ribosomal subunit maturation is controlled by a dual key lock.
Elife, 10, 2021
|
|
1EI0
 
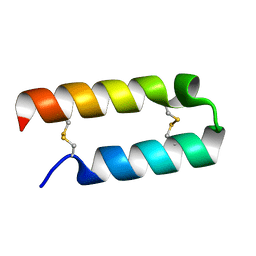 | |
1CXO
 
 | | REFINED THREE-DIMENSIONAL SOLUTION STRUCTURE OF A SNAKE CARDIOTOXIN: ANALYSIS OF THE SIDE-CHAIN ORGANISATION SUGGESTS THE EXISTENCE OF A POSSIBLE PHOSPHOLIPID BINDING SITE | | Descriptor: | CARDIOTOXIN GAMMA | | Authors: | Gilquin, B, Roumestand, C, Zinn-Justin, S, Menez, A, Toma, F. | | Deposit date: | 1994-11-07 | | Release date: | 1994-12-20 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Refined three-dimensional solution structure of a snake cardiotoxin: analysis of the side-chain organization suggests the existence of a possible phospholipid binding site.
Biopolymers, 33, 1993
|
|
6FNC
 
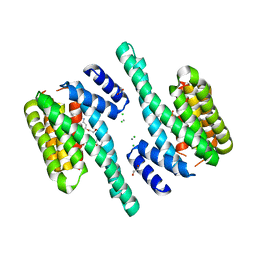 | |
1EII
 
 | | NMR STRUCTURE OF HOLO CELLULAR RETINOL-BINDING PROTEIN II | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN II, RETINOL | | Authors: | Lu, J, Lin, C.L, Tang, C, Ponder, J.W, Kao, J.L, Cistola, D.P, Li, E. | | Deposit date: | 2000-02-25 | | Release date: | 2000-08-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Binding of retinol induces changes in rat cellular retinol-binding protein II conformation and backbone dynamics.
J.Mol.Biol., 300, 2000
|
|
6RBE
 
 | | State 2 of yeast Tsr1-TAP Rps20-Deltaloop pre-40S particles | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Shayan, R, Mitterer, V, Ferreira-Cerca, S, Murat, G, Enne, T, Rinaldi, D, Weigl, S, Omanic, H, Gleizes, P.E, Kressler, D, Pertschy, B, Plisson-Chastang, C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism.
Nat Commun, 10, 2019
|
|
6RBD
 
 | | State 1 of yeast Tsr1-TAP Rps20-Deltaloop pre-40S particles | | Descriptor: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Shayan, R, Mitterer, V, Ferreira-Cerca, S, Murat, G, Enne, T, Rinaldi, D, Weigl, S, Omanic, H, Gleizes, P.E, Kressler, D, Pertschy, B, Plisson-Chastang, C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism.
Nat Commun, 10, 2019
|
|
7TWK
 
 | |
7TWM
 
 | |
