7F1P
 
 | | Crystal structure of Pseudomonas putida methionine gamma-lyase Q349S mutant ligand-free form. | | Descriptor: | L-methionine gamma-lyase | | Authors: | Okawa, A, Handa, H, Yasuda, E, Murota, M, Kudo, D, Tamura, T, Shiba, T, Inagaki, K. | | Deposit date: | 2021-06-09 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization and application of l-methionine gamma-lyase Q349S mutant enzyme with an enhanced activity toward l-homocysteine.
J.Biosci.Bioeng., 133, 2022
|
|
2DG7
 
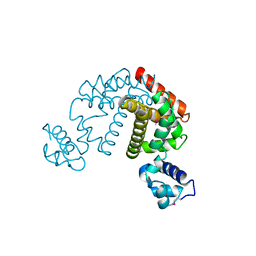 | | Crystal structure of the putative transcriptional regulator SCO0337 from Streptomyces coelicolor A3(2) | | Descriptor: | putative transcriptional regulator | | Authors: | Hayashi, T, Tanaka, Y, Sakai, N, Yao, M, Tamura, T, Tanaka, I. | | Deposit date: | 2006-03-08 | | Release date: | 2007-03-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the putative transcriptional regulator SCO0337 from Streptomyces coelicolor A3(2)
To be Published
|
|
2DG8
 
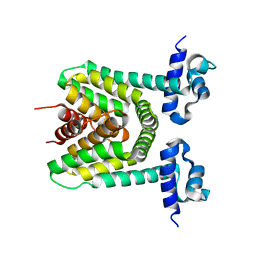 | | Crystal structure of the putative trasncriptional regulator SCO7518 from Streptomyces coelicolor A3(2) | | Descriptor: | putative tetR-family transcriptional regulatory protein | | Authors: | Hayashi, T, Watanabe, N, Sakai, N, Tamura, T, Yao, M, Tanaka, I. | | Deposit date: | 2006-03-08 | | Release date: | 2007-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of the putative transcriptional regulator SCO7518 from Streptomyces coelicolor A3(2)
To be Published
|
|
2DG6
 
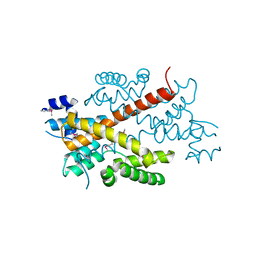 | | Crystal structure of the putative transcriptional regulator SCO5550 from Streptomyces coelicolor A3(2) | | Descriptor: | putative transcriptional regulator | | Authors: | Hayashi, T, Tanaka, Y, Sakai, N, Yao, M, Tamura, T, Tanaka, I. | | Deposit date: | 2006-03-08 | | Release date: | 2007-03-13 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and genomic DNA analysis of a putative transcription factor SCO5550 from Streptomyces coelicolor A3(2): regulating the expression of gene sco5551 as a transcriptional activator with a novel dimer shape
Biochem. Biophys. Res. Commun., 435, 2013
|
|
2D6Y
 
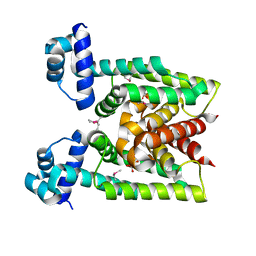 | | Crystal Structure of transcriptional factor SCO4008 from Streptomyces coelicolor A3(2) | | Descriptor: | L(+)-TARTARIC ACID, putative tetR family regulatory protein | | Authors: | Hayashi, T, Tanaka, Y, Sakai, N, Yao, M, Tamura, T, Tanaka, I. | | Deposit date: | 2005-11-15 | | Release date: | 2006-10-31 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SCO4008, a Putative TetR Transcriptional Repressor from Streptomyces coelicolor A3(2), Regulates Transcription of sco4007 by Multidrug Recognition.
J.Mol.Biol., 425, 2013
|
|
3WEC
 
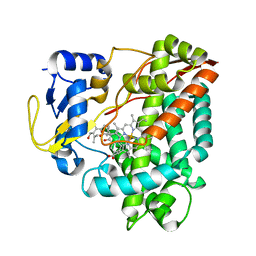 | | Structure of P450 RauA (CYP1050A1) complexed with a biosynthetic intermediate of aurachin RE | | Descriptor: | 3-[(2E,6E,9R)-9-hydroxy-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]-2-methylquinolin-4(1H)-one, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yasutake, Y, Kitagawa, W, Tamura, T. | | Deposit date: | 2013-07-03 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of the quinoline N-hydroxylating cytochrome P450 RauA, an essential enzyme that confers antibiotic activity on aurachin alkaloids
Febs Lett., 588, 2014
|
|
2D4V
 
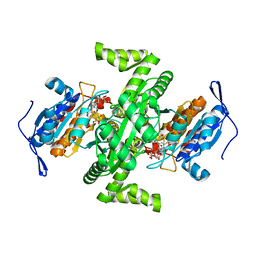 | | Crystal structure of NAD dependent isocitrate dehydrogenase from Acidithiobacillus thiooxidans | | Descriptor: | CITRATE ANION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, isocitrate dehydrogenase | | Authors: | Imada, K, Tamura, T, Namba, K, Inagaki, K. | | Deposit date: | 2005-10-24 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and quantum chemical analysis of NAD+-dependent isocitrate dehydrogenase: hydride transfer and co-factor specificity
Proteins, 70, 2008
|
|
2E1M
 
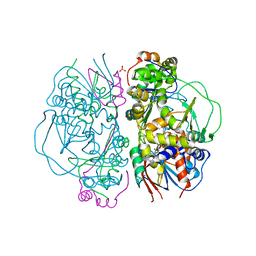 | | Crystal Structure of L-Glutamate Oxidase from Streptomyces sp. X-119-6 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-glutamate oxidase, PHOSPHATE ION | | Authors: | Sasaki, C, Kashima, A, Sakaguchi, C, Mizuno, H, Arima, J, Kusakabe, H, Tamura, T, Sugio, S, Inagaki, K. | | Deposit date: | 2006-10-26 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characterization of l-glutamate oxidase from Streptomyces sp. X-119-6
Febs J., 276, 2009
|
|
2D4W
 
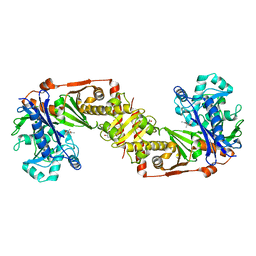 | |
3VK3
 
 | | Crystal Structure of L-Methionine gamma-Lyase from Pseudomonas putida C116H Mutant Complexed with L-methionine | | Descriptor: | METHIONINE, Methionine gamma-lyase | | Authors: | Fukumoto, M, Kudou, D, Murano, S, Shiba, T, Sato, D, Tamura, T, Harada, S, Inagaki, K. | | Deposit date: | 2011-11-07 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The role of amino acid residues in the active site of L-methionine gamma-lyase from Pseudomonas putida.
Biosci.Biotechnol.Biochem., 76, 2012
|
|
3VK4
 
 | | Crystal Structure of L-Methionine gamma-Lyase from Pseudomonas putida C116H Mutant complexed with L-homocysteine | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Methionine gamma-lyase | | Authors: | Fukumoto, M, Kudou, D, Murano, S, Shiba, T, Sato, D, Tamura, T, Harada, S, Inagaki, K. | | Deposit date: | 2011-11-07 | | Release date: | 2012-09-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The role of amino acid residues in the active site of L-methionine gamma-lyase from Pseudomonas putida.
Biosci.Biotechnol.Biochem., 76, 2012
|
|
3VTZ
 
 | |
3VRM
 
 | | Structure of cytochrome P450 Vdh mutant T107A with bound vitamin D3 | | Descriptor: | (1S,3Z)-3-[(2E)-2-[(1R,3AR,7AS)-7A-METHYL-1-[(2R)-6-METHYLHEPTAN-2-YL]-2,3,3A,5,6,7-HEXAHYDRO-1H-INDEN-4-YLIDENE]ETHYLI DENE]-4-METHYLIDENE-CYCLOHEXAN-1-OL, PROTOPORPHYRIN IX CONTAINING FE, Vitamin D(3) 25-hydroxylase | | Authors: | Nishioka, T, Yasutake, Y, Tamura, T. | | Deposit date: | 2012-04-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | A single mutation at the ferredoxin binding site of p450 vdh enables efficient biocatalytic production of 25-hydroxyvitamin d3.
Chembiochem, 14, 2013
|
|
3VK2
 
 | | Crystal Structure of L-Methionine gamma-Lyase from Pseudomonas putida C116H Mutant. | | Descriptor: | Methionine gamma-lyase, SULFATE ION | | Authors: | Fukumoto, M, Kudou, D, Murano, S, Shiba, T, Sato, D, Tamura, T, Harada, S, Inagaki, K. | | Deposit date: | 2011-11-07 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The role of amino acid residues in the active site of L-methionine gamma-lyase from Pseudomonas putida.
Biosci.Biotechnol.Biochem., 76, 2012
|
|
2Z36
 
 | | Crystal structure of cytochrome P450 MoxA from Nonomuraea recticatena (CYP105) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cytochrome P450 type compactin 3'',4''-hydroxylase, FE (III) ION, ... | | Authors: | Yasutake, Y, Fujii, Y, Fujii, T, Arisawa, A, Tamura, T. | | Deposit date: | 2007-06-02 | | Release date: | 2007-08-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of cytochrome P450 MoxA from Nonomuraea recticatena (CYP105)
Biochem.Biophys.Res.Commun., 361, 2007
|
|
3AUU
 
 | | Crystal structure of Bacillus megaterium glucose dehydrogenase 4 in complex with D-glucose | | Descriptor: | Glucose 1-dehydrogenase 4, beta-D-glucopyranose | | Authors: | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, T. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided mutagenesis for the improvement of substrate specificity of Bacillus megaterium glucose 1-dehydrogenase IV
Febs J., 279, 2012
|
|
3AUT
 
 | | Crystal structure of Bacillus megaterium glucose dehydrogenase 4 in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glucose 1-dehydrogenase 4 | | Authors: | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, T. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided mutagenesis for the improvement of substrate specificity of Bacillus megaterium glucose 1-dehydrogenase IV
Febs J., 279, 2012
|
|
3AUS
 
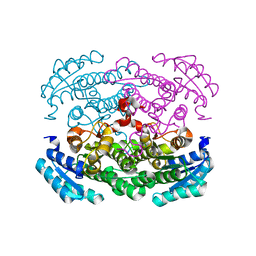 | |
3AY6
 
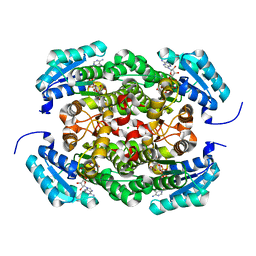 | | Crystal structure of Bacillus megaterium glucose dehydrogenase 4 A258F mutant in complex with NADH and D-glucose | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, Glucose 1-dehydrogenase 4, ... | | Authors: | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, T. | | Deposit date: | 2011-04-29 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-guided mutagenesis for the improvement of substrate specificity of Bacillus megaterium glucose 1-dehydrogenase IV
Febs J., 279, 2012
|
|
3AY7
 
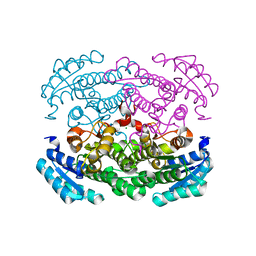 | | Crystal structure of Bacillus megaterium glucose dehydrogenase 4 G259A mutant | | Descriptor: | CHLORIDE ION, Glucose 1-dehydrogenase 4 | | Authors: | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, T. | | Deposit date: | 2011-04-29 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided mutagenesis for the improvement of substrate specificity of Bacillus megaterium glucose 1-dehydrogenase IV
Febs J., 279, 2012
|
|
3A51
 
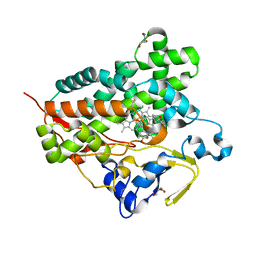 | | Structure of cytochrome P450 Vdh mutant (Vdh-K1) obtained by directed evolution with bound 25-hydroxyvitamin D3 | | Descriptor: | 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yasutake, Y, Fujii, Y, Cheon, W.K, Arisawa, A, Tamura, T. | | Deposit date: | 2009-07-24 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for enhancement of sequential vitamin D3 hydroxylation activities by directed evolution of cytochrome P450 vitamin D3 hydroxylase
J.Biol.Chem., 285, 2010
|
|
3A4H
 
 | | Structure of cytochrome P450 vdh from Pseudonocardia autotrophica (orthorhombic crystal form) | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Vitamin D hydroxylase | | Authors: | Yasutake, Y, Fujii, Y, Cheon, W.K, Arisawa, A, Tamura, T. | | Deposit date: | 2009-07-07 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural evidence for enhancement of sequential vitamin D3 hydroxylation activities by directed evolution of cytochrome P450 vitamin D3 hydroxylase
J.Biol.Chem., 285, 2010
|
|
3A4G
 
 | | Structure of cytochrome P450 vdh from Pseudonocardia autotrophica (trigonal crystal form) | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, PROTOPORPHYRIN IX CONTAINING FE, Vitamin D hydroxylase | | Authors: | Yasutake, Y, Fujii, Y, Cheon, W.K, Arisawa, A, Tamura, T. | | Deposit date: | 2009-07-07 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural evidence for enhancement of sequential vitamin D3 hydroxylation activities by directed evolution of cytochrome P450 vitamin D3 hydroxylase
J.Biol.Chem., 285, 2010
|
|
3A4Z
 
 | | Structure of cytochrome P450 Vdh mutant (Vdh-K1) obtained by directed evolution | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Yasutake, Y, Fujii, Y, Cheon, W.K, Arisawa, A, Tamura, T. | | Deposit date: | 2009-07-24 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural evidence for enhancement of sequential vitamin D3 hydroxylation activities by directed evolution of cytochrome P450 vitamin D3 hydroxylase
J.Biol.Chem., 285, 2010
|
|
3A50
 
 | | Structure of cytochrome P450 Vdh mutant (Vdh-K1) obtained by directed evolution with bound vitamin D3 | | Descriptor: | (1S,3Z)-3-[(2E)-2-[(1R,3AR,7AS)-7A-METHYL-1-[(2R)-6-METHYLHEPTAN-2-YL]-2,3,3A,5,6,7-HEXAHYDRO-1H-INDEN-4-YLIDENE]ETHYLI DENE]-4-METHYLIDENE-CYCLOHEXAN-1-OL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yasutake, Y, Fujii, Y, Cheon, W.K, Arisawa, A, Tamura, T. | | Deposit date: | 2009-07-24 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural evidence for enhancement of sequential vitamin D3 hydroxylation activities by directed evolution of cytochrome P450 vitamin D3 hydroxylase
J.Biol.Chem., 285, 2010
|
|
